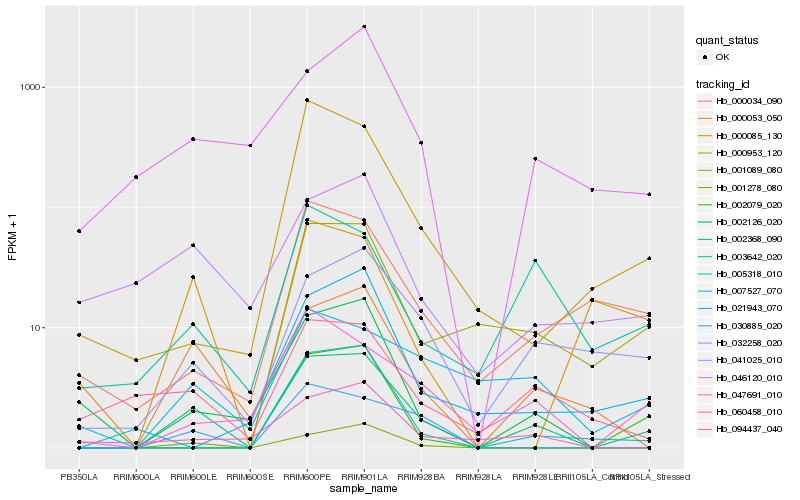
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002368_090 |
0.0 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Populus euphratica] |
| 2 |
Hb_000053_050 |
0.2116684709 |
- |
- |
hypothetical protein F775_01878 [Aegilops tauschii] |
| 3 |
Hb_001278_080 |
0.2328306791 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein JCGZ_09311 [Jatropha curcas] |
| 4 |
Hb_032258_020 |
0.2464791154 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Malus domestica] |
| 5 |
Hb_030885_020 |
0.2551335734 |
- |
- |
- |
| 6 |
Hb_047691_010 |
0.2622187511 |
- |
- |
Uncharacterized protein F383_01974 [Gossypium arboreum] |
| 7 |
Hb_002126_020 |
0.2637915264 |
- |
- |
ferrochelatase [Medicago truncatula] |
| 8 |
Hb_094437_040 |
0.2753229094 |
- |
- |
ankyrin repeat family protein, partial [Manihot esculenta] |
| 9 |
Hb_003642_020 |
0.2802040119 |
- |
- |
chlorophyll(ide) b reductase, partial [Pyrus x bretschneideri] |
| 10 |
Hb_046120_010 |
0.2823897339 |
- |
- |
- |
| 11 |
Hb_000953_120 |
0.2834498122 |
- |
- |
- |
| 12 |
Hb_000034_090 |
0.2839126174 |
transcription factor |
TF Family: M-type |
MPF2-like, partial [Dunalia fasciculata] |
| 13 |
Hb_007527_070 |
0.2890020787 |
- |
- |
PREDICTED: uncharacterized protein LOC105637197 [Jatropha curcas] |
| 14 |
Hb_005318_010 |
0.2905724549 |
- |
- |
hypothetical protein POPTR_0012s15190g, partial [Populus trichocarpa] |
| 15 |
Hb_060458_010 |
0.2908936277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_041025_010 |
0.2911853487 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 17 |
Hb_021943_070 |
0.2924646424 |
- |
- |
- |
| 18 |
Hb_002079_020 |
0.2936288396 |
- |
- |
- |
| 19 |
Hb_001089_080 |
0.2945791413 |
- |
- |
hypothetical protein glysoja_044047 [Glycine soja] |
| 20 |
Hb_000085_130 |
0.2952642649 |
- |
- |
- |