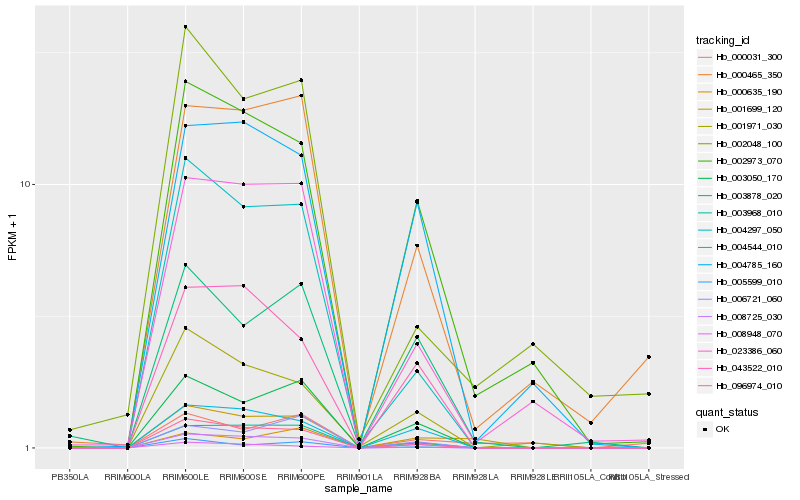
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001699_120 |
0.0 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 2 |
Hb_002973_070 |
0.1830364195 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 3 |
Hb_008725_030 |
0.1868602727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_096974_010 |
0.195096947 |
- |
- |
PREDICTED: uncharacterized protein LOC105123384 isoform X1 [Populus euphratica] |
| 5 |
Hb_003968_010 |
0.1972462098 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_003050_170 |
0.2011478341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004297_050 |
0.202119457 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_023386_060 |
0.2130868178 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 9 |
Hb_004544_010 |
0.2138617811 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 10 |
Hb_003878_020 |
0.215870982 |
- |
- |
Epidermis-specific secreted glycoprotein EP1 precursor, putative [Ricinus communis] |
| 11 |
Hb_001971_030 |
0.2206187013 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 12 |
Hb_043522_010 |
0.2208672792 |
- |
- |
- |
| 13 |
Hb_000465_350 |
0.2229192589 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2 [Jatropha curcas] |
| 14 |
Hb_004785_160 |
0.2252329338 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 15 |
Hb_002048_100 |
0.226620866 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Jatropha curcas] |
| 16 |
Hb_000031_300 |
0.2313355776 |
- |
- |
PREDICTED: histidine kinase 5 [Jatropha curcas] |
| 17 |
Hb_008948_070 |
0.2320569791 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_005599_010 |
0.2340944072 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 19 |
Hb_000635_190 |
0.2347307754 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103322127 [Prunus mume] |
| 20 |
Hb_006721_060 |
0.2348816349 |
- |
- |
RnaseH (mitochondrion) [Malus domestica] |