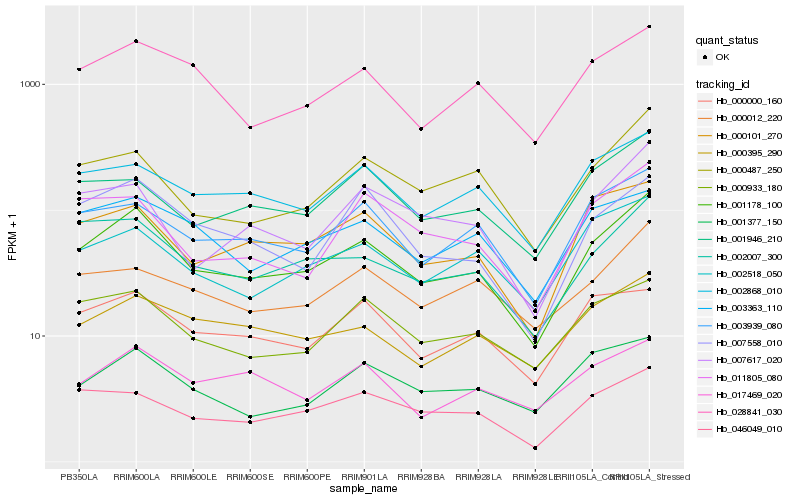
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001178_100 |
0.0 |
- |
- |
60S acidic ribosomal protein P2-4 [Theobroma cacao] |
| 2 |
Hb_002007_300 |
0.1012033461 |
- |
- |
hypothetical protein JCGZ_23450 [Jatropha curcas] |
| 3 |
Hb_003939_080 |
0.1077614217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_028841_030 |
0.1083713727 |
- |
- |
- |
| 5 |
Hb_000101_270 |
0.1088566443 |
- |
- |
pleckstrin homology domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_000487_250 |
0.1113385228 |
- |
- |
60S ribosomal protein L27e, putative [Ricinus communis] |
| 7 |
Hb_002868_010 |
0.11177057 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 8 |
Hb_046049_010 |
0.1138176228 |
- |
- |
PREDICTED: far upstream element-binding protein 2 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000933_180 |
0.1143410523 |
- |
- |
PREDICTED: chorismate mutase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002518_050 |
0.1150155746 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM10 [Jatropha curcas] |
| 11 |
Hb_000395_290 |
0.1160272523 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-8 [Jatropha curcas] |
| 12 |
Hb_001377_150 |
0.1189387472 |
- |
- |
hypothetical protein JCGZ_14266 [Jatropha curcas] |
| 13 |
Hb_001946_210 |
0.1193086488 |
- |
- |
ribosomal protein l7ae, putative [Ricinus communis] |
| 14 |
Hb_007558_010 |
0.1196510279 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM9-2 [Jatropha curcas] |
| 15 |
Hb_007617_020 |
0.1200606169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003363_110 |
0.1200788309 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_000012_220 |
0.1221316713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000000_160 |
0.1241927949 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 19 |
Hb_011805_080 |
0.1243391305 |
- |
- |
PREDICTED: uncharacterized protein LOC100242357 [Vitis vinifera] |
| 20 |
Hb_017469_020 |
0.125988742 |
- |
- |
- |