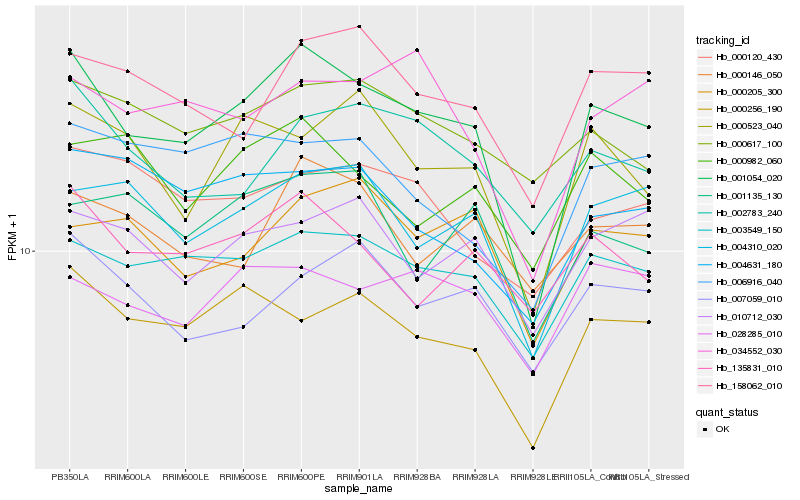
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001054_020 |
0.0 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 2 |
Hb_003549_150 |
0.0942122703 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 3 |
Hb_000256_190 |
0.0953838925 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 4 |
Hb_004310_020 |
0.1026244068 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000523_040 |
0.1034232457 |
- |
- |
PREDICTED: auxin response factor 8 isoform X1 [Jatropha curcas] |
| 6 |
Hb_158062_010 |
0.1077250754 |
- |
- |
PREDICTED: uncharacterized protein LOC105634413 [Jatropha curcas] |
| 7 |
Hb_135831_010 |
0.1111792602 |
- |
- |
hypothetical protein JCGZ_13374 [Jatropha curcas] |
| 8 |
Hb_002783_240 |
0.1114188643 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 9 |
Hb_034552_030 |
0.1117024878 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 10 |
Hb_028285_010 |
0.1137709775 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 11 |
Hb_000205_300 |
0.1150392949 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 12 |
Hb_004631_180 |
0.1153942458 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 13 |
Hb_006916_040 |
0.1158065226 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 14 |
Hb_007059_010 |
0.1159894996 |
- |
- |
PREDICTED: uncharacterized protein LOC105642418 [Jatropha curcas] |
| 15 |
Hb_001135_130 |
0.1164963574 |
- |
- |
hypothetical protein CISIN_1g0171752mg, partial [Citrus sinensis] |
| 16 |
Hb_000120_430 |
0.1172043804 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 17 |
Hb_000982_060 |
0.1175860731 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 18 |
Hb_010712_030 |
0.1176279427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000617_100 |
0.1181596611 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 20 |
Hb_000146_050 |
0.1207505832 |
- |
- |
PREDICTED: protein unc-50 homolog isoform X2 [Gossypium raimondii] |