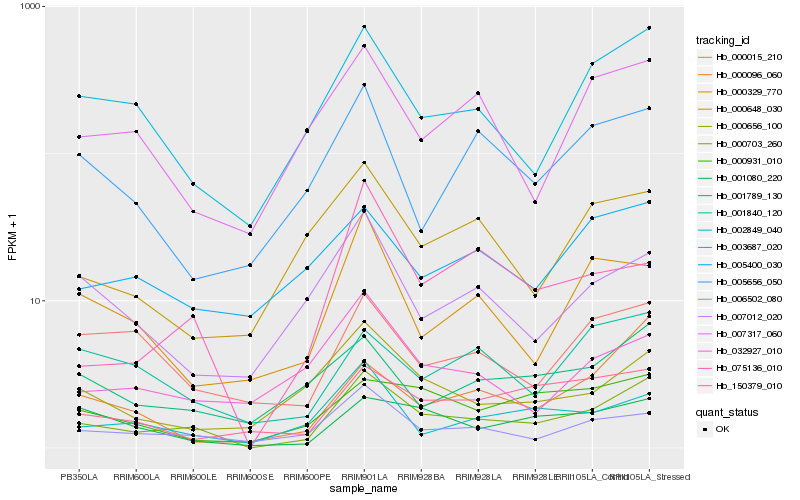
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_260 |
0.0 |
- |
- |
N-acetylglucosamine kinase, putative [Ricinus communis] |
| 2 |
Hb_003687_020 |
0.1766858 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 3 |
Hb_006502_080 |
0.1803048389 |
- |
- |
- |
| 4 |
Hb_000656_100 |
0.202762502 |
- |
- |
PREDICTED: uncharacterized protein LOC105633651 [Jatropha curcas] |
| 5 |
Hb_001789_130 |
0.2043770214 |
- |
- |
short-chain dehydrogenase Tic32 family protein [Populus trichocarpa] |
| 6 |
Hb_002849_040 |
0.2044333198 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 7 |
Hb_150379_010 |
0.2074970395 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 8 |
Hb_001080_220 |
0.2082986761 |
- |
- |
TPA: hypothetical protein ZEAMMB73_966926 [Zea mays] |
| 9 |
Hb_000931_010 |
0.2111496484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007317_060 |
0.2115328128 |
- |
- |
60S ribosomal protein L10aB [Hevea brasiliensis] |
| 11 |
Hb_000015_210 |
0.2152340302 |
- |
- |
PREDICTED: uncharacterized protein LOC105647781 [Jatropha curcas] |
| 12 |
Hb_000096_060 |
0.2156012804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007012_020 |
0.216023141 |
- |
- |
hypothetical protein POPTR_0005s11940g [Populus trichocarpa] |
| 14 |
Hb_000648_030 |
0.2180162289 |
- |
- |
hypothetical protein PHAVU_002G055100g [Phaseolus vulgaris] |
| 15 |
Hb_075136_010 |
0.2183678506 |
- |
- |
Protein SSU72, putative [Ricinus communis] |
| 16 |
Hb_005400_030 |
0.2189847207 |
- |
- |
PREDICTED: LYR motif-containing protein At3g19508 [Jatropha curcas] |
| 17 |
Hb_005656_050 |
0.2203044461 |
- |
- |
PREDICTED: protein FAM32A-like [Jatropha curcas] |
| 18 |
Hb_001840_120 |
0.2204791188 |
- |
- |
PREDICTED: uncharacterized protein LOC104228746 [Nicotiana sylvestris] |
| 19 |
Hb_032927_010 |
0.2207686924 |
- |
- |
- |
| 20 |
Hb_000329_770 |
0.221472067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |