| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
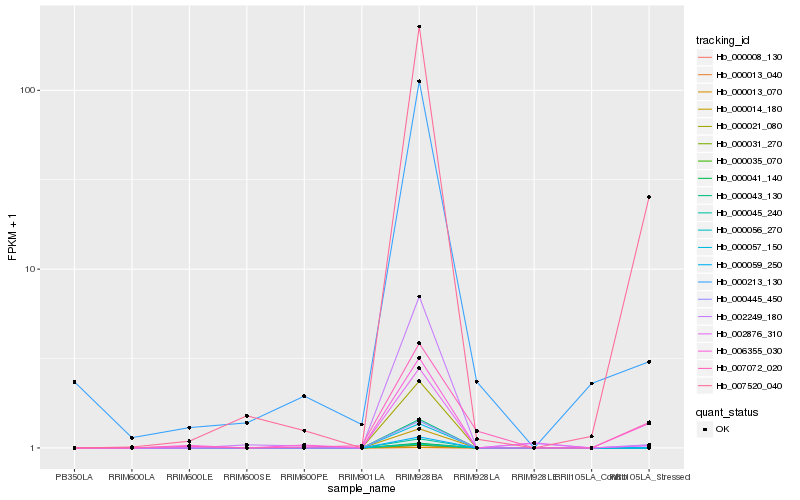
Hb_000445_450 |
0.0 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_007520_040 |
0.0487257321 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 3 |
Hb_006355_030 |
0.1374435509 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |
| 4 |
Hb_002876_310 |
0.1943595504 |
- |
- |
hypothetical protein JCGZ_03912 [Jatropha curcas] |
| 5 |
Hb_002249_180 |
0.2124641557 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000213_130 |
0.2132821505 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 7 |
Hb_007072_020 |
0.2149193807 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 8 |
Hb_000008_130 |
0.2162160813 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000013_040 |
0.2162160813 |
- |
- |
- |
| 10 |
Hb_000013_070 |
0.2162160813 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_000014_180 |
0.2162160813 |
- |
- |
PREDICTED: endo-1,4-beta-xylanase D-like [Fragaria vesca subsp. vesca] |
| 12 |
Hb_000021_080 |
0.2162160813 |
- |
- |
- |
| 13 |
Hb_000031_270 |
0.2162160813 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000035_070 |
0.2162160813 |
- |
- |
PREDICTED: uncharacterized protein LOC104814513 [Tarenaya hassleriana] |
| 15 |
Hb_000041_140 |
0.2162160813 |
- |
- |
- |
| 16 |
Hb_000043_130 |
0.2162160813 |
- |
- |
- |
| 17 |
Hb_000045_240 |
0.2162160813 |
- |
- |
- |
| 18 |
Hb_000056_270 |
0.2162160813 |
- |
- |
Origin recognition complex subunit, putative [Ricinus communis] |
| 19 |
Hb_000057_150 |
0.2162160813 |
- |
- |
hypothetical protein EUTSA_v10009202mg [Eutrema salsugineum] |
| 20 |
Hb_000059_250 |
0.2162160813 |
- |
- |
PREDICTED: 40S ribosomal protein S8 [Jatropha curcas] |