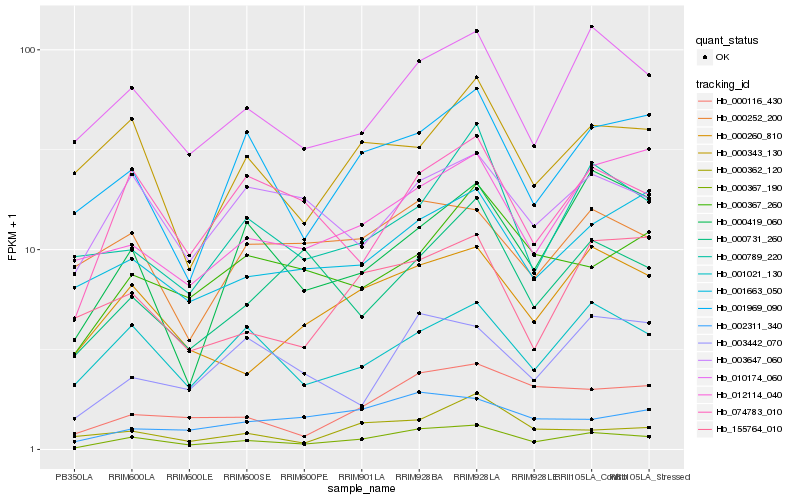
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_190 |
0.0 |
- |
- |
hypothetical protein RCOM_1470550 [Ricinus communis] |
| 2 |
Hb_010174_060 |
0.1345402674 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_000260_810 |
0.1379949709 |
- |
- |
- |
| 4 |
Hb_001021_130 |
0.1437209084 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 5 |
Hb_000116_430 |
0.1448239715 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_074783_010 |
0.147243796 |
- |
- |
PREDICTED: uncharacterized protein LOC105649970 [Jatropha curcas] |
| 7 |
Hb_001969_090 |
0.1500696966 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 8 |
Hb_002311_340 |
0.1574890218 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 9 |
Hb_001663_050 |
0.1589924755 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |
| 10 |
Hb_003647_060 |
0.1605024651 |
- |
- |
PREDICTED: uncharacterized protein LOC105646679 isoform X2 [Jatropha curcas] |
| 11 |
Hb_012114_040 |
0.1642322016 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000419_060 |
0.1646086542 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 13 |
Hb_000789_220 |
0.1656265545 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 14 |
Hb_155764_010 |
0.1665152834 |
- |
- |
- |
| 15 |
Hb_000367_260 |
0.1689449893 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 16 |
Hb_000731_260 |
0.1691424626 |
- |
- |
hypothetical protein RCOM_0905090 [Ricinus communis] |
| 17 |
Hb_003442_070 |
0.1691510805 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 18 |
Hb_000362_120 |
0.1703005293 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 19 |
Hb_000343_130 |
0.1706763392 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |
| 20 |
Hb_000252_200 |
0.1712289697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |