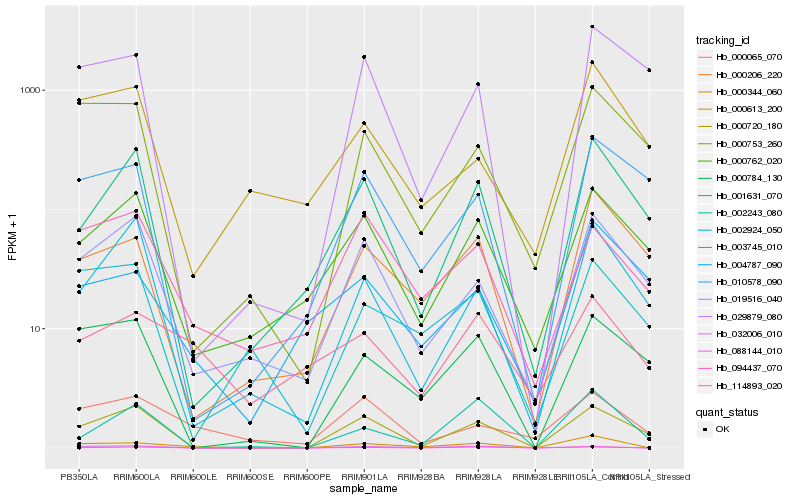
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000344_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 2 |
Hb_088144_010 |
0.2392821648 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 3 |
Hb_000206_220 |
0.2670468591 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 9 [Jatropha curcas] |
| 4 |
Hb_000720_180 |
0.2807992552 |
- |
- |
- |
| 5 |
Hb_019516_040 |
0.2877031172 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 6 |
Hb_032006_010 |
0.2877676854 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 7 |
Hb_002243_080 |
0.2893342758 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Vitis vinifera] |
| 8 |
Hb_000784_130 |
0.2899232486 |
- |
- |
hypothetical protein JCGZ_22813 [Jatropha curcas] |
| 9 |
Hb_000753_260 |
0.2934139267 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 10 |
Hb_000065_070 |
0.2940311298 |
- |
- |
hypothetical protein CISIN_1g039077mg [Citrus sinensis] |
| 11 |
Hb_001631_070 |
0.2963273138 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 12 |
Hb_004787_090 |
0.2990240035 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 13 |
Hb_094437_070 |
0.3014130424 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 14 |
Hb_000762_020 |
0.3016513204 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 15 |
Hb_002924_050 |
0.303668916 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 16 |
Hb_000613_200 |
0.3052299347 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 17 |
Hb_003745_010 |
0.3072621271 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 18 |
Hb_029879_080 |
0.3074056596 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_114893_020 |
0.3079186845 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_010578_090 |
0.3079784489 |
- |
- |
HEV2.1 [Hevea brasiliensis] |