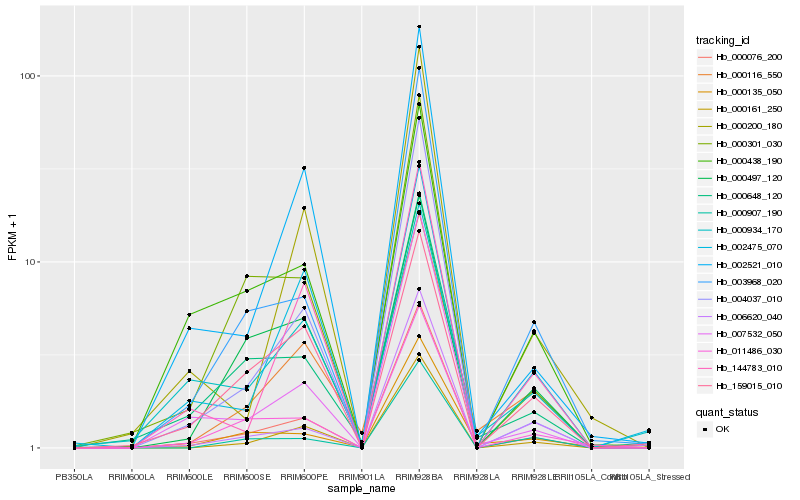
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_550 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000161_250 |
0.1177588111 |
transcription factor |
TF Family: Tify |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000076_200 |
0.1391871727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000497_120 |
0.1489461175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_144783_010 |
0.1503941963 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 6 |
Hb_000648_120 |
0.156041613 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 7 |
Hb_000907_190 |
0.1565670023 |
- |
- |
PREDICTED: uncharacterized protein At4g15970-like [Jatropha curcas] |
| 8 |
Hb_000135_050 |
0.1568373225 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like protein kinase At2g19210-like isoform X2 [Citrus sinensis] |
| 9 |
Hb_002475_070 |
0.1625591816 |
- |
- |
hypothetical protein POPTR_0017s02440g, partial [Populus trichocarpa] |
| 10 |
Hb_011486_030 |
0.1625890678 |
- |
- |
branched-chain amino acid aminotransferase, putative [Ricinus communis] |
| 11 |
Hb_002521_010 |
0.163701194 |
- |
- |
cinnamate 4-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_003968_020 |
0.1672247993 |
- |
- |
PREDICTED: limonoid UDP-glucosyltransferase-like [Glycine max] |
| 13 |
Hb_000200_180 |
0.1701851859 |
- |
- |
- |
| 14 |
Hb_159015_010 |
0.1702606002 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000934_170 |
0.1710915945 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_006620_040 |
0.1756233549 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 17 |
Hb_007532_050 |
0.1760406864 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 18 |
Hb_000301_030 |
0.1774886123 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 19 |
Hb_004037_010 |
0.1787263968 |
- |
- |
PREDICTED: uncharacterized protein LOC105632369 [Jatropha curcas] |
| 20 |
Hb_000438_190 |
0.1801933887 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |