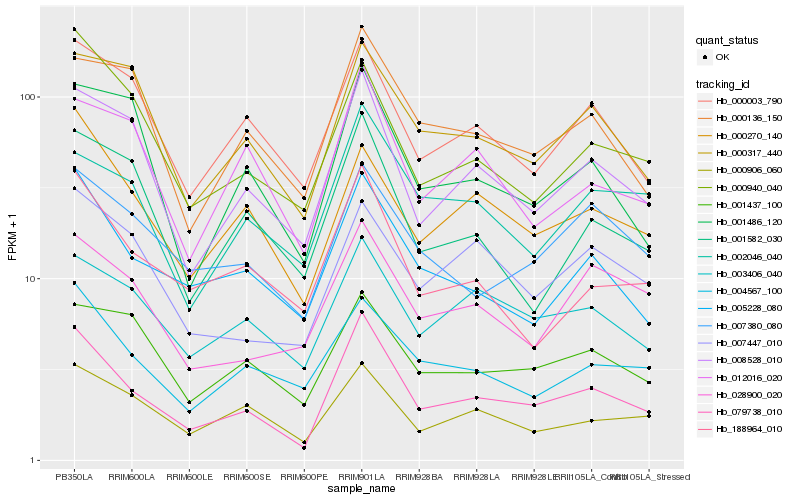
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079738_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g036582mg, partial [Citrus sinensis] |
| 2 |
Hb_008528_010 |
0.1201094241 |
- |
- |
PREDICTED: uncharacterized protein LOC105628498 [Jatropha curcas] |
| 3 |
Hb_005228_080 |
0.1336944092 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-I [Jatropha curcas] |
| 4 |
Hb_188964_010 |
0.1425821557 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 5 |
Hb_000003_790 |
0.1443509576 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 6 |
Hb_000906_060 |
0.1448553937 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g08820 [Populus euphratica] |
| 7 |
Hb_000270_140 |
0.1448957086 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003406_040 |
0.146845928 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 9 |
Hb_000940_040 |
0.1487493126 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 10 |
Hb_004567_100 |
0.1504325393 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 11 |
Hb_012016_020 |
0.1505234208 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105636265 [Jatropha curcas] |
| 12 |
Hb_001486_120 |
0.150568963 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 13 |
Hb_000136_150 |
0.1509845893 |
- |
- |
hypothetical protein VITISV_038366 [Vitis vinifera] |
| 14 |
Hb_000317_440 |
0.1516387665 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001582_030 |
0.1531130885 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 16 |
Hb_007380_080 |
0.1542567362 |
transcription factor |
TF Family: G2-like |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 17 |
Hb_028900_020 |
0.1554313105 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_007447_010 |
0.1566266073 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB3 isoform X3 [Jatropha curcas] |
| 19 |
Hb_002046_040 |
0.1571464197 |
- |
- |
PREDICTED: topless-related protein 4 [Malus domestica] |
| 20 |
Hb_001437_100 |
0.1578690053 |
- |
- |
PREDICTED: DNA-binding protein REB1-like [Jatropha curcas] |