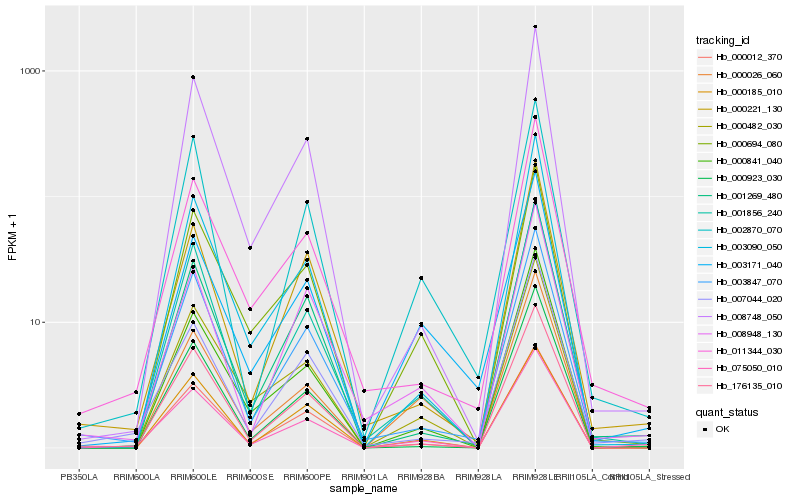
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075050_010 |
0.0 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_001856_240 |
0.0891037739 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 3 |
Hb_000482_030 |
0.0908385287 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |
| 4 |
Hb_000012_370 |
0.0937320304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_176135_010 |
0.097435657 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_003847_070 |
0.1019949211 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000694_080 |
0.1023081129 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 8 |
Hb_008748_050 |
0.102314156 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_003090_050 |
0.1061635445 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 10 |
Hb_001269_480 |
0.1065176239 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_007044_020 |
0.1067715904 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_002870_070 |
0.107055773 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 13 |
Hb_000221_130 |
0.1085663579 |
- |
- |
thylakoid lumenal 29 kDa protein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003171_040 |
0.1095651036 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 15 |
Hb_000841_040 |
0.1101526807 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000923_030 |
0.1104546751 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 17 |
Hb_011344_030 |
0.1115574903 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_008948_130 |
0.1146495332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000185_010 |
0.1149671421 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 20 |
Hb_000026_060 |
0.1149887598 |
- |
- |
beta-1,3-glucanase [Manihot esculenta] |