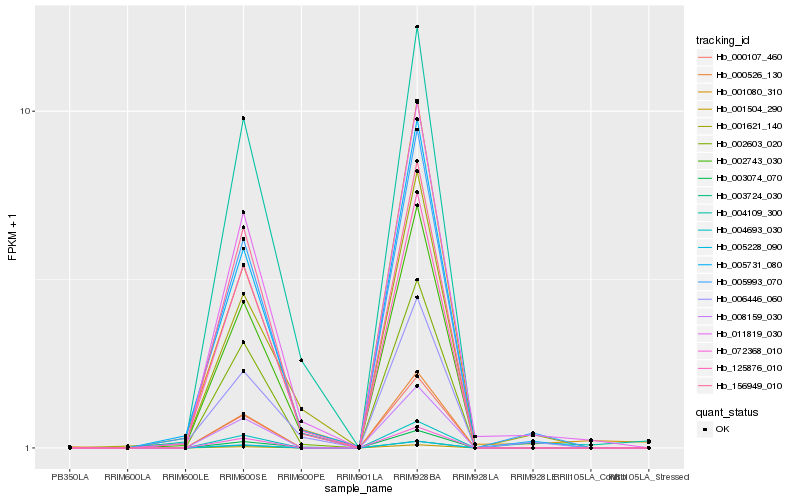
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011819_030 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_005993_070 |
0.0919957548 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 3 |
Hb_002743_030 |
0.0976522173 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 4 |
Hb_002603_020 |
0.0992858141 |
- |
- |
PREDICTED: O-acetyl-ADP-ribose deacetylase MACROD2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_004109_300 |
0.1026318884 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 6 |
Hb_005731_080 |
0.1032903612 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 7 |
Hb_003074_070 |
0.1072467318 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_125876_010 |
0.1101375968 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_156949_010 |
0.1114645254 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 10 |
Hb_001621_140 |
0.1185822023 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 11 |
Hb_001080_310 |
0.1197589761 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006446_060 |
0.1204003543 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 13 |
Hb_008159_030 |
0.1237329251 |
- |
- |
hypothetical protein B456_004G088300 [Gossypium raimondii] |
| 14 |
Hb_000107_460 |
0.1238380707 |
- |
- |
PREDICTED: uncharacterized protein LOC104585930 [Nelumbo nucifera] |
| 15 |
Hb_001504_290 |
0.1244244656 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 16 |
Hb_072368_010 |
0.1245064758 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 17 |
Hb_003724_030 |
0.1245980671 |
- |
- |
- |
| 18 |
Hb_004693_030 |
0.124844924 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 19 |
Hb_000526_130 |
0.1248524442 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 20 |
Hb_005228_090 |
0.1252473158 |
- |
- |
PREDICTED: magnesium transporter MRS2-3-like [Citrus sinensis] |