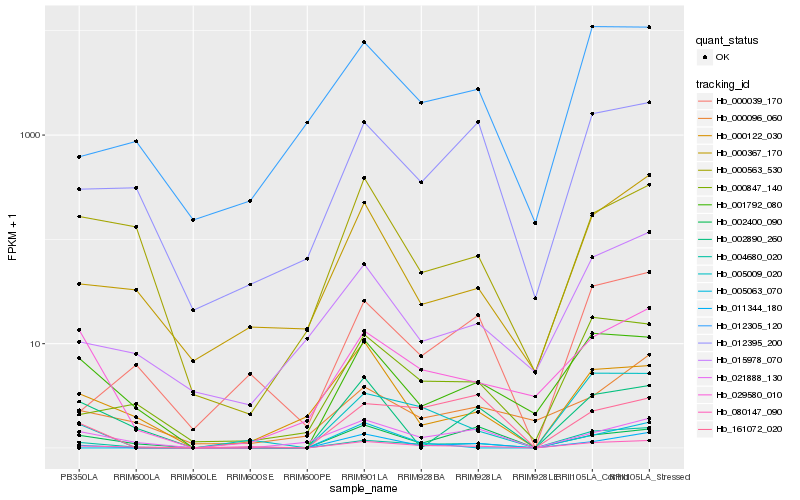
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_180 |
0.0 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 2 |
Hb_000367_170 |
0.2399008417 |
- |
- |
PREDICTED: LYR motif-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_080147_090 |
0.24830056 |
- |
- |
hypothetical protein JCGZ_07271 [Jatropha curcas] |
| 4 |
Hb_021888_130 |
0.2526980444 |
- |
- |
- |
| 5 |
Hb_012395_200 |
0.2608086877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002400_090 |
0.2642669596 |
- |
- |
- |
| 7 |
Hb_002890_260 |
0.2650261199 |
- |
- |
- |
| 8 |
Hb_004680_020 |
0.2658676109 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105789475 [Gossypium raimondii] |
| 9 |
Hb_015978_070 |
0.266992191 |
- |
- |
- |
| 10 |
Hb_000122_030 |
0.2687831364 |
- |
- |
PREDICTED: uncharacterized protein LOC104606081, partial [Nelumbo nucifera] |
| 11 |
Hb_005009_020 |
0.2700540398 |
- |
- |
- |
| 12 |
Hb_001792_080 |
0.2729906429 |
- |
- |
unknown [Populus trichocarpa] |
| 13 |
Hb_000847_140 |
0.2740281174 |
- |
- |
- |
| 14 |
Hb_161072_020 |
0.2791480817 |
- |
- |
- |
| 15 |
Hb_029580_010 |
0.27916387 |
- |
- |
PREDICTED: uncharacterized protein LOC105633393 [Jatropha curcas] |
| 16 |
Hb_005063_070 |
0.2793775958 |
- |
- |
PREDICTED: PRA1 family protein E [Jatropha curcas] |
| 17 |
Hb_000563_530 |
0.2815276622 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |
| 18 |
Hb_012305_120 |
0.2815438055 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 19 |
Hb_000039_170 |
0.2825276371 |
- |
- |
protein MOTHER of FT and TF 1-like [Jatropha curcas] |
| 20 |
Hb_000096_060 |
0.2832102654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |