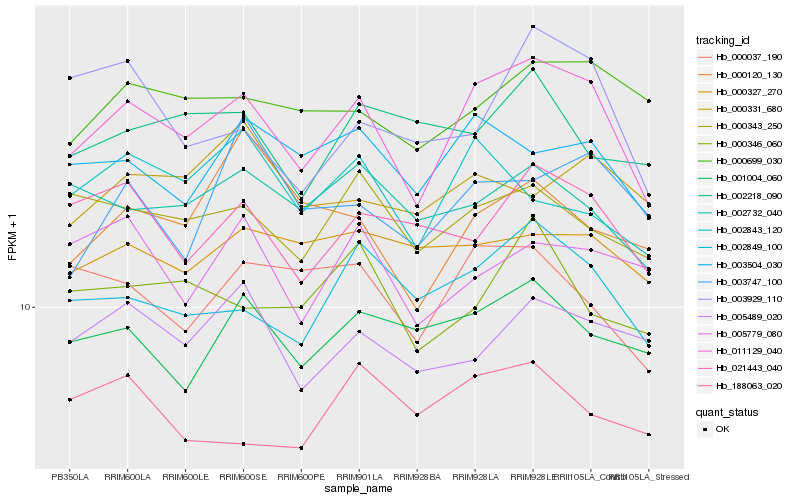
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011129_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645602 [Jatropha curcas] |
| 2 |
Hb_002849_100 |
0.0745389935 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_003504_030 |
0.0796223586 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000343_250 |
0.0831646634 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 5 |
Hb_000037_190 |
0.0851097643 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 6 |
Hb_000699_030 |
0.0856274619 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_021443_040 |
0.0866730795 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 8 |
Hb_002732_040 |
0.0871086793 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001004_060 |
0.0881509816 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 10 |
Hb_005489_020 |
0.089238113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002843_120 |
0.0914333445 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000346_060 |
0.0917427869 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 13 |
Hb_000327_270 |
0.093439771 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_005779_080 |
0.0949909584 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003929_110 |
0.0962395382 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_188063_020 |
0.0973974939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003747_100 |
0.097875779 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000120_130 |
0.0980160767 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 19 |
Hb_002218_090 |
0.0980352235 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000331_680 |
0.0985724596 |
- |
- |
Protein bem46, putative [Ricinus communis] |