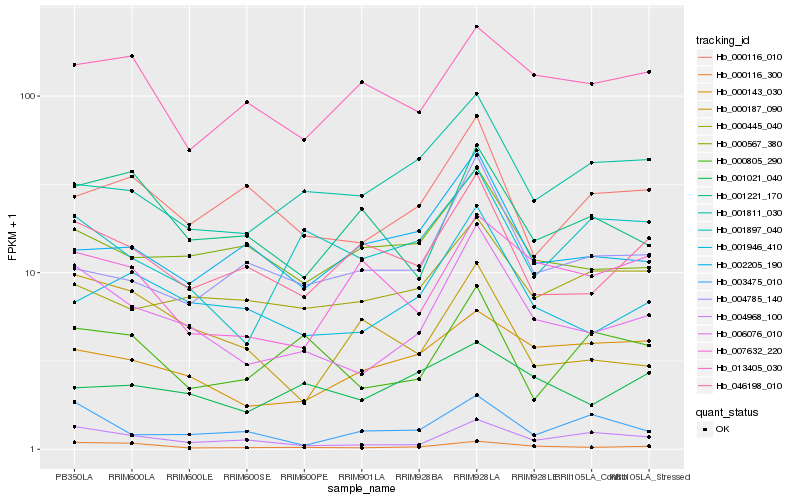
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006076_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 2 |
Hb_000116_300 |
0.1316007244 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_004968_100 |
0.1483175511 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 4 |
Hb_001946_410 |
0.1499697265 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_046198_010 |
0.1662763697 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 6 |
Hb_000567_380 |
0.1704923364 |
- |
- |
ubiquitin specific protease 39 and snrnp assembly factor, putative [Ricinus communis] |
| 7 |
Hb_000116_010 |
0.1841454471 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000445_040 |
0.185469199 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000143_030 |
0.1862574622 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 10 |
Hb_004785_140 |
0.1876598951 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000805_290 |
0.1892121115 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 12 |
Hb_001221_170 |
0.1894335039 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50390, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001811_030 |
0.1897970932 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 14 |
Hb_001897_040 |
0.1912066633 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 15 |
Hb_002205_190 |
0.1922005612 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 16 |
Hb_003475_010 |
0.1948626803 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 17 |
Hb_000187_090 |
0.1963646742 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 18 |
Hb_001021_040 |
0.1966401819 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 19 |
Hb_007632_220 |
0.1976946916 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 20 |
Hb_013405_030 |
0.1990421487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |