| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
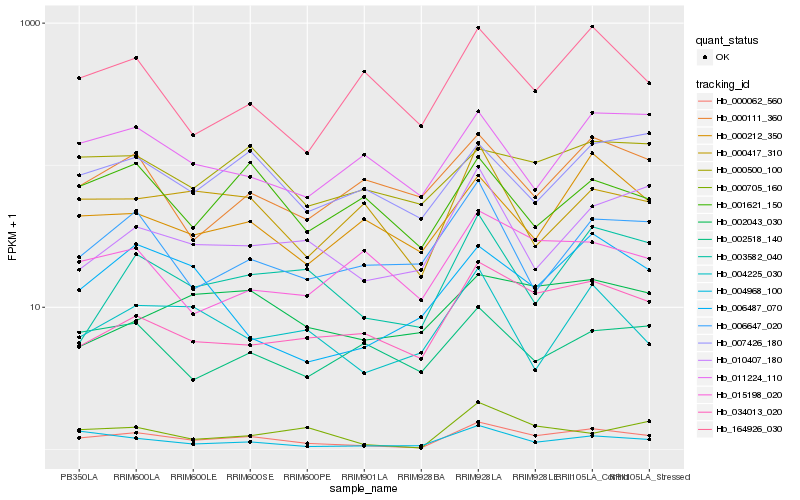
Hb_000062_560 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_034013_020 |
0.1582904411 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 3 |
Hb_004968_100 |
0.1640389738 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 4 |
Hb_000500_100 |
0.1782589217 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |
| 5 |
Hb_000705_160 |
0.1793289628 |
- |
- |
hypothetical protein CISIN_1g042743mg [Citrus sinensis] |
| 6 |
Hb_003582_040 |
0.1810607154 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0010s11160g [Populus trichocarpa] |
| 7 |
Hb_000417_310 |
0.1823009578 |
- |
- |
uncharacterized protein LOC100272531 [Zea mays] |
| 8 |
Hb_007426_180 |
0.18456721 |
- |
- |
PREDICTED: probable inositol transporter 2 [Jatropha curcas] |
| 9 |
Hb_000212_350 |
0.1868213822 |
- |
- |
glucosamine-fructose-6-phosphate aminotransferase, putative [Ricinus communis] |
| 10 |
Hb_164926_030 |
0.1881576094 |
- |
- |
PREDICTED: uncharacterized protein LOC105645602 [Jatropha curcas] |
| 11 |
Hb_006487_070 |
0.1897304876 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 14 [Jatropha curcas] |
| 12 |
Hb_011224_110 |
0.1911675245 |
- |
- |
Pre-mRNA-splicing factor cwc15, putative [Ricinus communis] |
| 13 |
Hb_002518_140 |
0.1914994861 |
- |
- |
hypothetical protein JCGZ_18870 [Jatropha curcas] |
| 14 |
Hb_010407_180 |
0.1919091526 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 15 |
Hb_006647_020 |
0.1919522641 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 16 |
Hb_001621_150 |
0.1920077659 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Jatropha curcas] |
| 17 |
Hb_002043_030 |
0.193792758 |
- |
- |
hypothetical protein RCOM_1486170 [Ricinus communis] |
| 18 |
Hb_015198_020 |
0.1952033223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000111_360 |
0.1956119683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004225_030 |
0.1959583552 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |