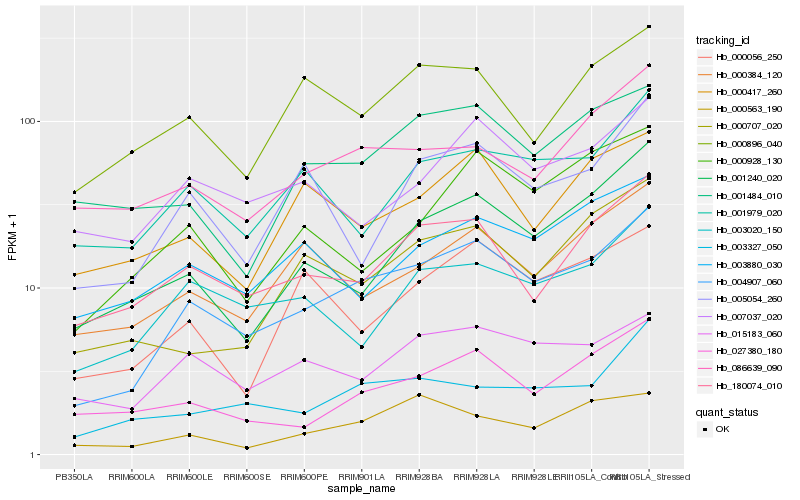
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004907_060 |
0.0 |
- |
- |
PREDICTED: tropinone reductase homolog At5g06060-like [Jatropha curcas] |
| 2 |
Hb_180074_010 |
0.1376543323 |
- |
- |
Single-stranded DNA-binding protein, mitochondrial precursor, putative [Ricinus communis] |
| 3 |
Hb_003327_050 |
0.1420667147 |
- |
- |
surfeit locus protein, putative [Ricinus communis] |
| 4 |
Hb_001240_020 |
0.1444190479 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 5 |
Hb_003020_150 |
0.1470155778 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 6 |
Hb_015183_060 |
0.1477756017 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 7 |
Hb_027380_180 |
0.1484497075 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 8 |
Hb_000384_120 |
0.1499196199 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001979_020 |
0.1505913784 |
- |
- |
hypothetical protein OsJ_31823 [Oryza sativa Japonica Group] |
| 10 |
Hb_000056_250 |
0.1524232582 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 11 |
Hb_000707_020 |
0.1530311096 |
- |
- |
hypothetical protein B456_002G255500 [Gossypium raimondii] |
| 12 |
Hb_007037_020 |
0.1531727882 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 13 |
Hb_000896_040 |
0.154970775 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 14 |
Hb_005054_260 |
0.155603577 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 15 |
Hb_086639_090 |
0.1557309043 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 16 |
Hb_001484_010 |
0.1558342605 |
- |
- |
hypothetical protein POPTR_0005s07440g [Populus trichocarpa] |
| 17 |
Hb_003880_030 |
0.1561041646 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000928_130 |
0.1561937225 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_000417_260 |
0.1571255991 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_000563_190 |
0.1574153812 |
- |
- |
hypothetical protein PRUPE_ppa002828mg [Prunus persica] |