| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
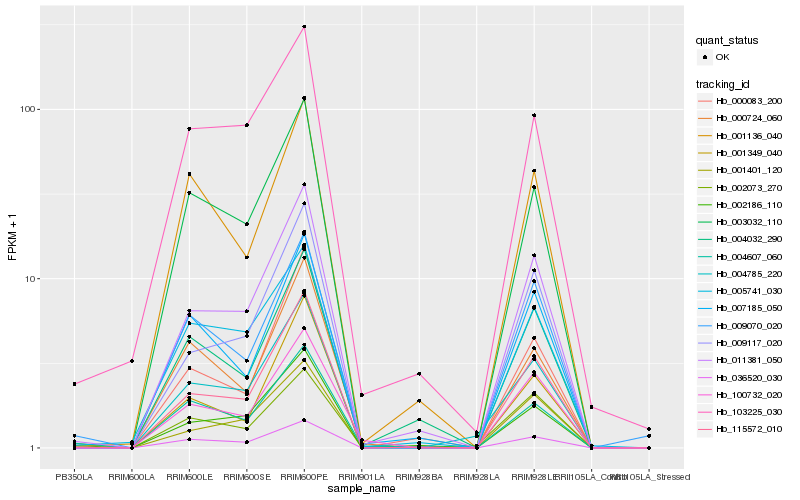
Hb_004785_220 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.2 [Populus euphratica] |
| 2 |
Hb_003032_110 |
0.0993416067 |
- |
- |
- |
| 3 |
Hb_115572_010 |
0.1025375895 |
- |
- |
PREDICTED: beta-galactosidase 6 [Jatropha curcas] |
| 4 |
Hb_011381_050 |
0.1099523001 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 5 |
Hb_002073_270 |
0.1217282076 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 6 |
Hb_036520_030 |
0.1242497609 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 7 |
Hb_009117_020 |
0.1253561238 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 8 |
Hb_005741_030 |
0.1306055139 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 9 |
Hb_002186_110 |
0.1310836037 |
- |
- |
hypothetical protein CICLE_v10024045mg, partial [Citrus clementina] |
| 10 |
Hb_001401_120 |
0.1315987618 |
- |
- |
hypothetical protein JCGZ_04273 [Jatropha curcas] |
| 11 |
Hb_103225_030 |
0.132893461 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_100732_020 |
0.1361612492 |
- |
- |
hypothetical protein POPTR_0001s15930g [Populus trichocarpa] |
| 13 |
Hb_000724_060 |
0.1396363998 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 14 |
Hb_000083_200 |
0.1410381928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007185_050 |
0.1415911407 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 16 |
Hb_001349_040 |
0.1419725893 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 17 |
Hb_001136_040 |
0.1424061019 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_009070_020 |
0.1427086577 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |
| 19 |
Hb_004032_290 |
0.1474487178 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_004607_060 |
0.1494216395 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |