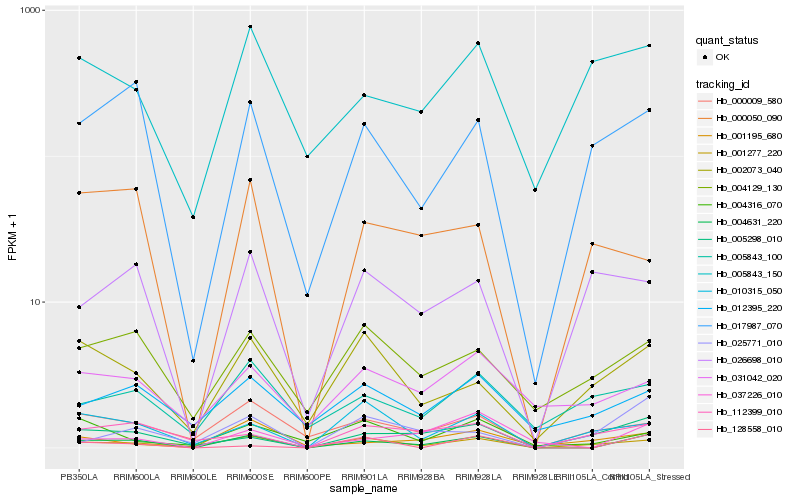
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004631_220 |
0.0 |
- |
- |
- |
| 2 |
Hb_001277_220 |
0.2365188955 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 3 |
Hb_037226_010 |
0.2640522318 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_017987_070 |
0.2667007506 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT22-like [Jatropha curcas] |
| 5 |
Hb_005298_010 |
0.2888778923 |
- |
- |
- |
| 6 |
Hb_004129_130 |
0.2950196311 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000009_580 |
0.2951907992 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 8 |
Hb_002073_040 |
0.2967875844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_012395_220 |
0.3009517972 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 10 |
Hb_010315_050 |
0.3017131002 |
- |
- |
hypothetical protein JCGZ_25247 [Jatropha curcas] |
| 11 |
Hb_128558_010 |
0.3052549056 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_005843_100 |
0.3084141436 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350-like [Jatropha curcas] |
| 13 |
Hb_112399_010 |
0.3094422826 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_001195_680 |
0.310441638 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 15 |
Hb_005843_150 |
0.3105245645 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 16 |
Hb_031042_020 |
0.3108363147 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g47460 [Jatropha curcas] |
| 17 |
Hb_000050_090 |
0.3110521949 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 18 |
Hb_026698_010 |
0.3117424277 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 19 |
Hb_004316_070 |
0.3128301577 |
- |
- |
E3 ubiquitin-protein ligase UPL5 -like protein [Gossypium arboreum] |
| 20 |
Hb_025771_010 |
0.3133402478 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |