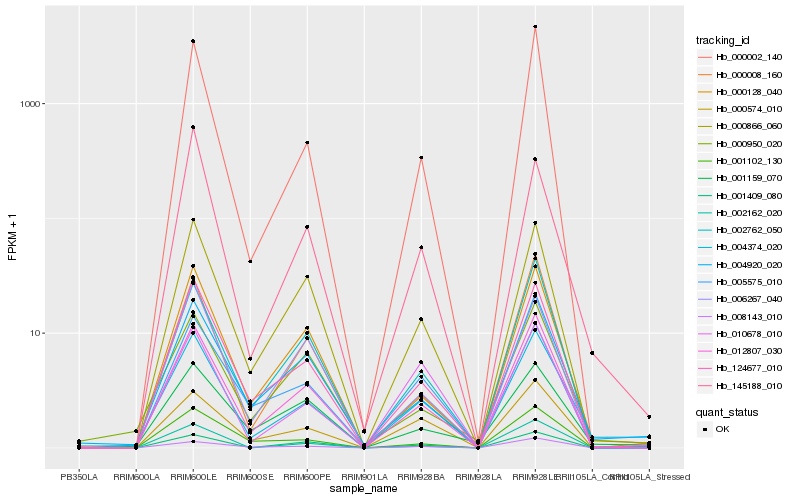
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004374_020 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2-like [Jatropha curcas] |
| 2 |
Hb_012807_030 |
0.112094269 |
- |
- |
PREDICTED: uncharacterized protein LOC105628042 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004920_020 |
0.126055993 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 4 |
Hb_124677_010 |
0.1309889481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000574_010 |
0.1409115961 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_06096 [Jatropha curcas] |
| 6 |
Hb_000002_140 |
0.143203587 |
- |
- |
hypothetical protein JCGZ_02318 [Jatropha curcas] |
| 7 |
Hb_000866_060 |
0.146151378 |
- |
- |
putative glycerol-3-phosphate acyltransferase [Vernicia fordii] |
| 8 |
Hb_000128_040 |
0.1506858587 |
- |
- |
PREDICTED: uncharacterized protein LOC105641228 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002762_050 |
0.1516451592 |
- |
- |
PREDICTED: uncharacterized protein LOC105640359 [Jatropha curcas] |
| 10 |
Hb_001102_130 |
0.1536077726 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 11 |
Hb_001409_080 |
0.1578184593 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Populus euphratica] |
| 12 |
Hb_008143_010 |
0.1578983574 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 13 |
Hb_005575_010 |
0.158050026 |
- |
- |
hypothetical protein POPTR_0004s02560g [Populus trichocarpa] |
| 14 |
Hb_001159_070 |
0.1584029278 |
- |
- |
DNA photolyase, putative [Ricinus communis] |
| 15 |
Hb_010678_010 |
0.1626190453 |
- |
- |
PREDICTED: alpha-glucosidase-like [Jatropha curcas] |
| 16 |
Hb_145188_010 |
0.1644043992 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 17 |
Hb_000950_020 |
0.1662674734 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 18 |
Hb_000008_160 |
0.1683167087 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 19 |
Hb_002162_020 |
0.169023286 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 20 |
Hb_006267_040 |
0.171545696 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |