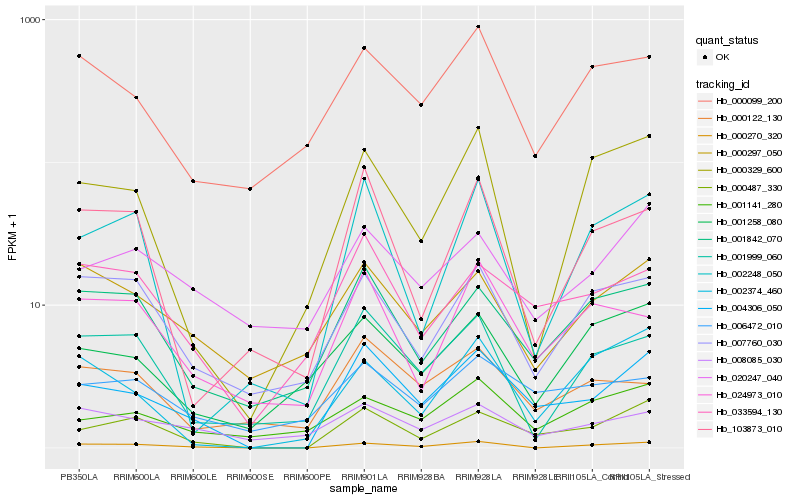
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004306_050 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000487_330 |
0.1277227628 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Populus euphratica] |
| 3 |
Hb_033594_130 |
0.1862759394 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Jatropha curcas] |
| 4 |
Hb_000270_320 |
0.1962096239 |
- |
- |
hypothetical protein RCOM_1512920 [Ricinus communis] |
| 5 |
Hb_001842_070 |
0.1964535299 |
- |
- |
PREDICTED: uncharacterized protein LOC105637106 [Jatropha curcas] |
| 6 |
Hb_006472_010 |
0.2104961459 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 7 |
Hb_020247_040 |
0.2126071466 |
- |
- |
PREDICTED: uncharacterized protein LOC105646910 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000099_200 |
0.2150790396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_024973_010 |
0.2164925241 |
- |
- |
PREDICTED: uncharacterized protein LOC105642097 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000297_050 |
0.2166587978 |
- |
- |
hypothetical protein JCGZ_08840 [Jatropha curcas] |
| 11 |
Hb_103873_010 |
0.2172997185 |
- |
- |
hypothetical protein POPTR_0008s14260g [Populus trichocarpa] |
| 12 |
Hb_001141_280 |
0.2175050539 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Vitis vinifera] |
| 13 |
Hb_002248_050 |
0.2177443307 |
- |
- |
PREDICTED: uncharacterized protein LOC105635697 [Jatropha curcas] |
| 14 |
Hb_000122_130 |
0.2185088372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000329_600 |
0.2189815592 |
- |
- |
PREDICTED: cell number regulator 6-like isoform X2 [Populus euphratica] |
| 16 |
Hb_002374_460 |
0.2194109036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001258_080 |
0.219563056 |
- |
- |
PREDICTED: uncharacterized protein LOC105639921 [Jatropha curcas] |
| 18 |
Hb_001999_060 |
0.2202860291 |
- |
- |
hypothetical protein POPTR_0009s05000g [Populus trichocarpa] |
| 19 |
Hb_007760_030 |
0.2207379447 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_15623 [Jatropha curcas] |
| 20 |
Hb_008085_030 |
0.2223830683 |
- |
- |
PREDICTED: uncharacterized protein LOC105639414 [Jatropha curcas] |