| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
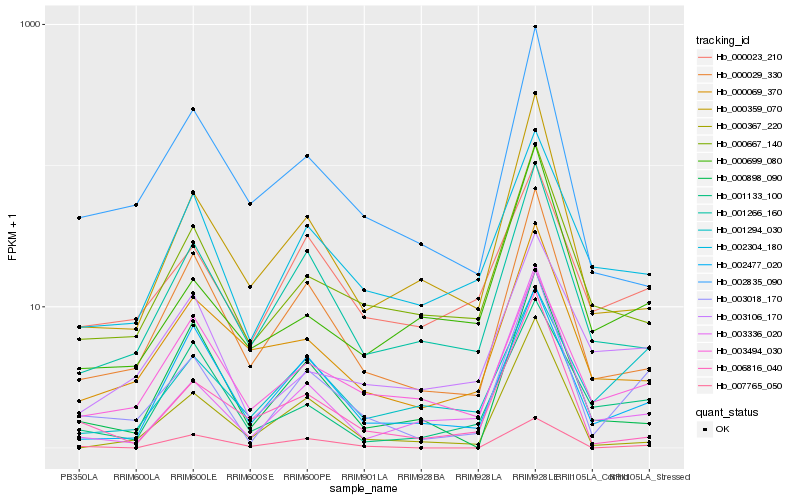
Hb_003018_170 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s11660g [Populus trichocarpa] |
| 2 |
Hb_000029_330 |
0.1789595887 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001266_160 |
0.1947274427 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000359_070 |
0.1955500037 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000667_140 |
0.2008604967 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003494_030 |
0.2031367017 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003336_020 |
0.2039466017 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS3, chloroplastic [Vitis vinifera] |
| 8 |
Hb_002304_180 |
0.2057164052 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlI, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000023_210 |
0.2092737545 |
- |
- |
PREDICTED: protein TIC 55, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002477_020 |
0.2100863783 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |
| 11 |
Hb_006816_040 |
0.2110026963 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 12 |
Hb_001294_030 |
0.2116071627 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |
| 13 |
Hb_000699_080 |
0.2127431343 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002835_090 |
0.2127553592 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 15 |
Hb_003106_170 |
0.2150989351 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 16 |
Hb_000069_370 |
0.2151298767 |
- |
- |
- |
| 17 |
Hb_001133_100 |
0.2156410053 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 18 |
Hb_000898_090 |
0.2188665853 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105633600 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007765_050 |
0.2192349408 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000367_220 |
0.2201452462 |
- |
- |
hypothetical protein RCOM_1470580 [Ricinus communis] |