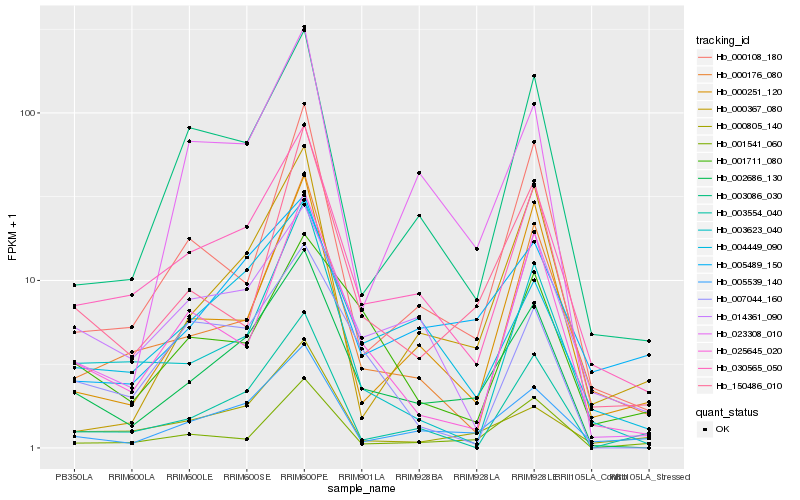
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_130 |
0.0 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105650401 [Jatropha curcas] |
| 2 |
Hb_030565_050 |
0.1404596336 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 3 |
Hb_005539_140 |
0.1513325177 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 4 |
Hb_004449_090 |
0.1672405793 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 5 |
Hb_150486_010 |
0.175257162 |
- |
- |
hypothetical protein CICLE_v100116733mg, partial [Citrus clementina] |
| 6 |
Hb_003086_030 |
0.1828387538 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 7 |
Hb_025645_020 |
0.1876530615 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 8 |
Hb_003623_040 |
0.1900421444 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_000176_080 |
0.1941447491 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 10 |
Hb_005489_150 |
0.1946156774 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 11 |
Hb_000108_180 |
0.1962735439 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 12 |
Hb_003554_040 |
0.1964253693 |
- |
- |
- |
| 13 |
Hb_000805_140 |
0.1976998281 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 14 |
Hb_001541_060 |
0.198157949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_023308_010 |
0.1997573987 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 16 |
Hb_014361_090 |
0.2001318635 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 17 |
Hb_007044_160 |
0.2005560934 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Populus euphratica] |
| 18 |
Hb_000251_120 |
0.2038307819 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001711_080 |
0.2038320001 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 20 |
Hb_000367_080 |
0.2038643586 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |