| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
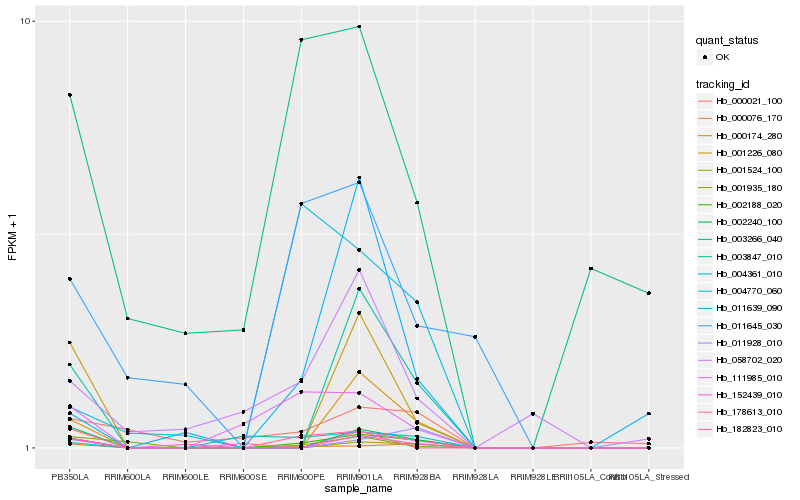
Hb_002188_020 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000076_170 |
0.2237143291 |
- |
- |
hypothetical protein JCGZ_00832 [Jatropha curcas] |
| 3 |
Hb_003847_010 |
0.2880145344 |
- |
- |
- |
| 4 |
Hb_002240_100 |
0.2887444852 |
- |
- |
- |
| 5 |
Hb_000174_280 |
0.300502506 |
- |
- |
- |
| 6 |
Hb_001524_100 |
0.3091878815 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 7 |
Hb_111985_010 |
0.3260668226 |
- |
- |
- |
| 8 |
Hb_001226_080 |
0.3305981023 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 9 |
Hb_152439_010 |
0.3448100648 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 10 |
Hb_182823_010 |
0.3577049668 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_011645_030 |
0.3663379315 |
- |
- |
- |
| 12 |
Hb_003266_040 |
0.370070098 |
- |
- |
PREDICTED: 6.7 kDa chloroplast outer envelope membrane protein-like [Jatropha curcas] |
| 13 |
Hb_058702_020 |
0.371253868 |
- |
- |
PREDICTED: uncharacterized protein LOC104604814 [Nelumbo nucifera] |
| 14 |
Hb_011928_010 |
0.3765140105 |
- |
- |
- |
| 15 |
Hb_001935_180 |
0.378215371 |
- |
- |
hypothetical protein VITISV_028082 [Vitis vinifera] |
| 16 |
Hb_004361_010 |
0.382956312 |
- |
- |
hypothetical protein CISIN_1g047084mg, partial [Citrus sinensis] |
| 17 |
Hb_000021_100 |
0.3831161742 |
- |
- |
- |
| 18 |
Hb_004770_060 |
0.3836659909 |
- |
- |
unnamed protein product, partial [Coffea canephora] |
| 19 |
Hb_178613_010 |
0.3852177566 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 20 |
Hb_011639_090 |
0.3869491473 |
- |
- |
PREDICTED: transcription factor bHLH75-like [Populus euphratica] |