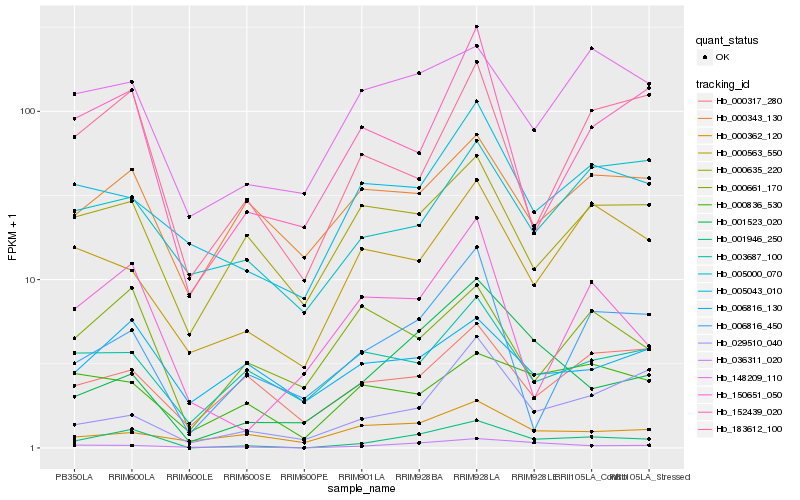
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_250 |
0.0 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 18 [Orycteropus afer afer] |
| 2 |
Hb_036311_020 |
0.2101106868 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_001523_020 |
0.2211381511 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 4 |
Hb_183612_100 |
0.2356391787 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 5 |
Hb_148209_110 |
0.2393680009 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 6 |
Hb_152439_020 |
0.2411022382 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_150651_050 |
0.246190638 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 8 |
Hb_005043_010 |
0.2469027151 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_000635_220 |
0.2505163909 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 10 |
Hb_006816_450 |
0.2523158563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000563_550 |
0.2529320395 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 12 |
Hb_000836_530 |
0.2553390318 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 13 |
Hb_029510_040 |
0.2556314609 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 14 |
Hb_000661_170 |
0.2565774882 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 15 |
Hb_005000_070 |
0.2577858615 |
- |
- |
hypothetical protein POPTR_0012s01650g [Populus trichocarpa] |
| 16 |
Hb_003687_100 |
0.2578272639 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Jatropha curcas] |
| 17 |
Hb_000317_280 |
0.2583314251 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At5g08490 [Jatropha curcas] |
| 18 |
Hb_000343_130 |
0.2591205851 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |
| 19 |
Hb_006816_130 |
0.2610566288 |
- |
- |
PREDICTED: uncharacterized protein LOC105647421 [Jatropha curcas] |
| 20 |
Hb_000362_120 |
0.2622675369 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |