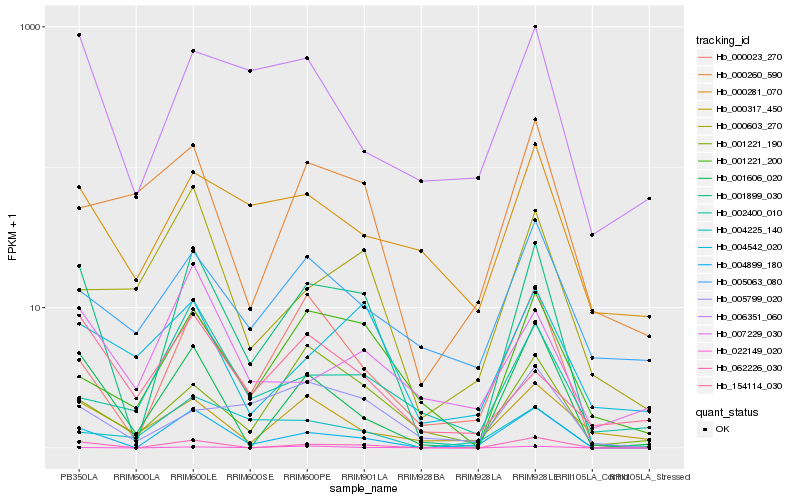
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001899_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 2 |
Hb_062226_030 |
0.1444181146 |
- |
- |
- |
| 3 |
Hb_004899_180 |
0.1573881123 |
- |
- |
hypothetical protein JCGZ_07531 [Jatropha curcas] |
| 4 |
Hb_001606_020 |
0.2120956219 |
- |
- |
PREDICTED: cell number regulator 2-like [Malus domestica] |
| 5 |
Hb_000023_270 |
0.2269630676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001221_200 |
0.2580911835 |
- |
- |
- |
| 7 |
Hb_001221_190 |
0.2770335723 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 8 |
Hb_006351_060 |
0.2816213029 |
- |
- |
PREDICTED: 17.8 kDa class I heat shock protein-like [Cucumis sativus] |
| 9 |
Hb_004542_020 |
0.2842920962 |
- |
- |
hypothetical chloroplast RF1 [Hevea brasiliensis] |
| 10 |
Hb_000281_070 |
0.2881260466 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 11 |
Hb_005799_020 |
0.288203787 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET6b [Jatropha curcas] |
| 12 |
Hb_000260_590 |
0.2884155724 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 13 |
Hb_022149_020 |
0.2885610667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000603_270 |
0.2891640595 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |
| 15 |
Hb_000317_450 |
0.2901523736 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004225_140 |
0.2942052128 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 17 |
Hb_005063_080 |
0.2949294548 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_007229_030 |
0.2954908716 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002400_010 |
0.3001721391 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 20 |
Hb_154114_030 |
0.3011372995 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |