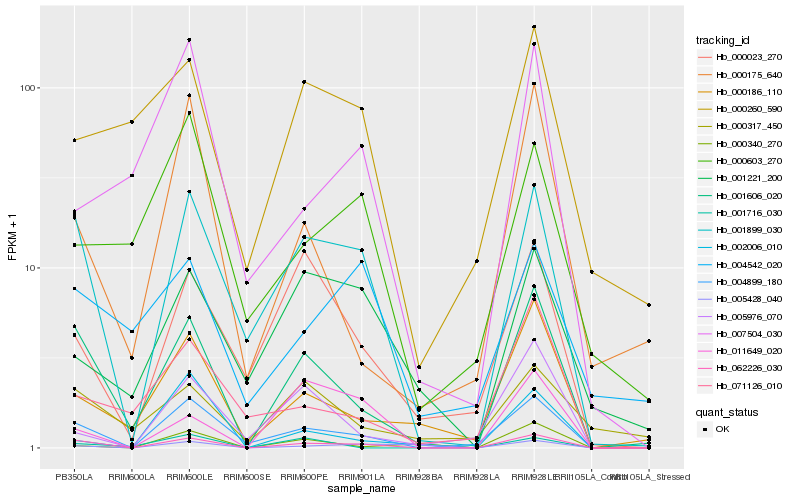
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_062226_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_001899_030 |
0.1444181146 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 3 |
Hb_001606_020 |
0.1450746439 |
- |
- |
PREDICTED: cell number regulator 2-like [Malus domestica] |
| 4 |
Hb_004899_180 |
0.1662568637 |
- |
- |
hypothetical protein JCGZ_07531 [Jatropha curcas] |
| 5 |
Hb_000186_110 |
0.2682289162 |
- |
- |
hypothetical protein CISIN_1g020639mg [Citrus sinensis] |
| 6 |
Hb_000023_270 |
0.2781284008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000175_640 |
0.2932162388 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005428_040 |
0.2950972573 |
- |
- |
- |
| 9 |
Hb_011649_020 |
0.2965941211 |
- |
- |
PREDICTED: uncharacterized protein LOC104212904 isoform X1 [Nicotiana sylvestris] |
| 10 |
Hb_071126_010 |
0.3023286891 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_005976_070 |
0.305503402 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 12 |
Hb_000260_590 |
0.3102264596 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 13 |
Hb_000317_450 |
0.3114739269 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002006_010 |
0.3135724683 |
- |
- |
Adenine nucleotide alpha hydrolases-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 15 |
Hb_004542_020 |
0.3172284849 |
- |
- |
hypothetical chloroplast RF1 [Hevea brasiliensis] |
| 16 |
Hb_001221_200 |
0.3174041678 |
- |
- |
- |
| 17 |
Hb_000340_270 |
0.3175524153 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 18 |
Hb_001716_030 |
0.3203183554 |
- |
- |
PREDICTED: uncharacterized protein LOC105633673 [Jatropha curcas] |
| 19 |
Hb_007504_030 |
0.3215978935 |
- |
- |
Rpl14, partial (chloroplast) [Schistostemon retusum] |
| 20 |
Hb_000603_270 |
0.3227343778 |
- |
- |
PREDICTED: uncharacterized protein LOC105638080 [Jatropha curcas] |