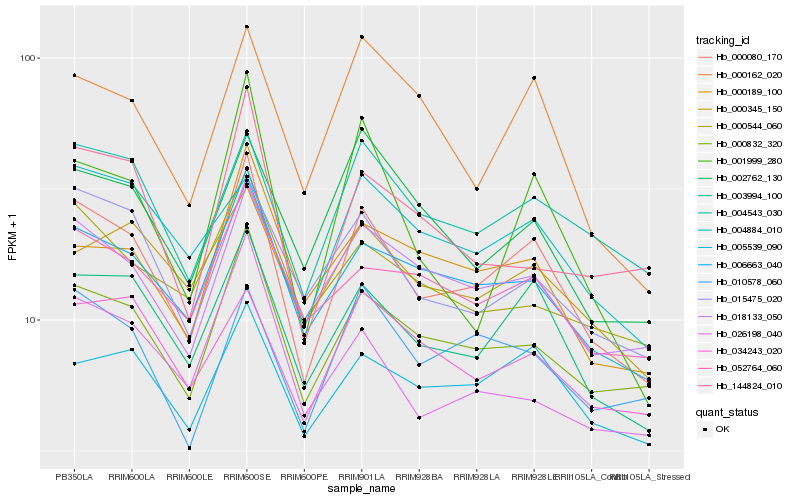
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000080_170 |
0.0 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_003994_100 |
0.0743595114 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000832_320 |
0.08777387 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_006663_040 |
0.0921309145 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004543_030 |
0.1016666007 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 6 |
Hb_018133_050 |
0.1016818877 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 7 |
Hb_015475_020 |
0.1066140311 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 8 |
Hb_005539_090 |
0.1123623361 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000162_020 |
0.1129436275 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 10 |
Hb_052764_060 |
0.1137862377 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 11 |
Hb_010578_060 |
0.1157729327 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 12 |
Hb_000189_100 |
0.1161404774 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 13 |
Hb_004884_010 |
0.1167027738 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC22 homolog [Jatropha curcas] |
| 14 |
Hb_002762_130 |
0.1175454674 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 15 |
Hb_000345_150 |
0.119460886 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001999_280 |
0.1201915853 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_034243_020 |
0.1213715197 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_144824_010 |
0.1216083319 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 19 |
Hb_026198_040 |
0.1216504708 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 20 |
Hb_000544_060 |
0.1223010797 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |