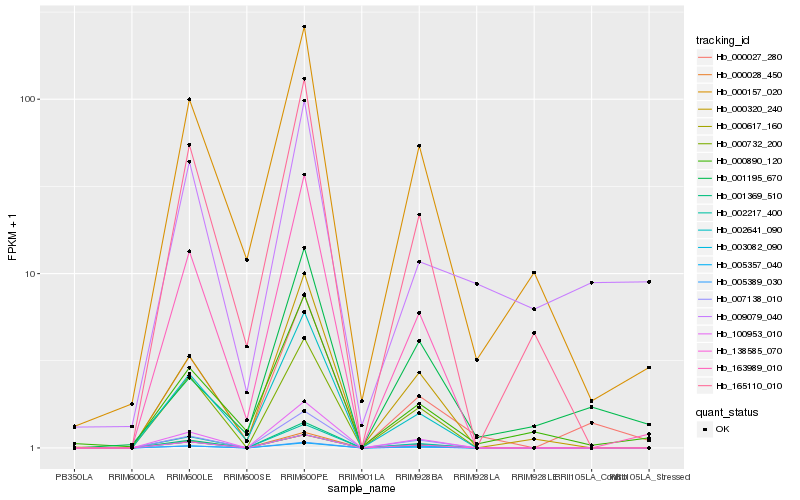
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642051 [Jatropha curcas] |
| 2 |
Hb_163989_010 |
0.1683446093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002641_090 |
0.1825316771 |
- |
- |
PREDICTED: uncharacterized protein LOC105643559 [Jatropha curcas] |
| 4 |
Hb_000157_020 |
0.1843274894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000890_120 |
0.1855891685 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009079_040 |
0.1881900026 |
- |
- |
BnaA02g30070D [Brassica napus] |
| 7 |
Hb_005389_030 |
0.1921642802 |
- |
- |
hypothetical protein VITISV_023348 [Vitis vinifera] |
| 8 |
Hb_000028_450 |
0.1939582508 |
- |
- |
PREDICTED: monoglyceride lipase-like [Jatropha curcas] |
| 9 |
Hb_005357_040 |
0.1940355764 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 10 |
Hb_002217_400 |
0.1954949182 |
- |
- |
polynucleotidyl transferase, ribonuclease H-like protein [Arabidopsis thaliana] |
| 11 |
Hb_003082_090 |
0.1963002319 |
- |
- |
hypothetical protein POPTR_0013s099451g, partial [Populus trichocarpa] |
| 12 |
Hb_100953_010 |
0.1965841256 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 13 |
Hb_138585_070 |
0.1982305612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001369_510 |
0.1985218988 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 15 |
Hb_007138_010 |
0.1990517942 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 16 |
Hb_000617_160 |
0.1993511964 |
- |
- |
- |
| 17 |
Hb_165110_010 |
0.2029623078 |
- |
- |
PREDICTED: kunitz-type elastase inhibitor BrEI-like [Jatropha curcas] |
| 18 |
Hb_000732_200 |
0.203399187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000320_240 |
0.2042940781 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 20 |
Hb_001195_670 |
0.2103568085 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |