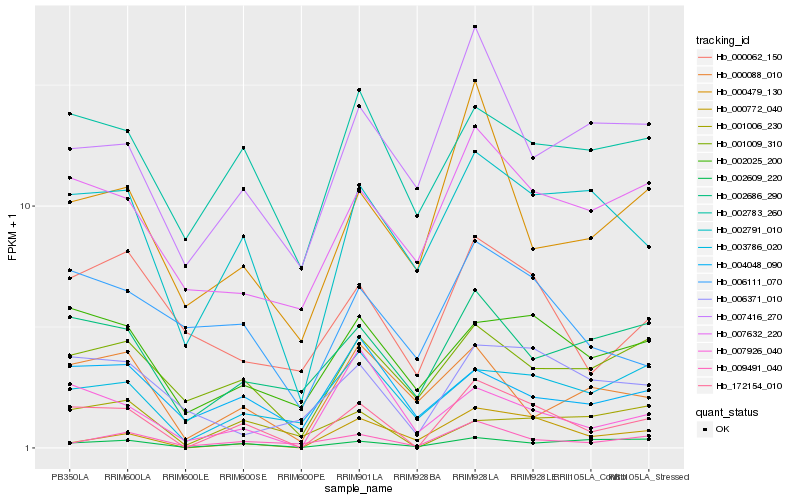
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172154_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_25247 [Jatropha curcas] |
| 2 |
Hb_002791_010 |
0.2405308518 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002609_220 |
0.2452986781 |
- |
- |
Leucine-rich repeat protein kinase family protein, putative [Theobroma cacao] |
| 4 |
Hb_009491_040 |
0.249522241 |
- |
- |
PREDICTED: uncharacterized protein LOC105636967 [Jatropha curcas] |
| 5 |
Hb_007926_040 |
0.2521155974 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 6 |
Hb_002025_200 |
0.2557133397 |
- |
- |
- |
| 7 |
Hb_000772_040 |
0.2578756648 |
- |
- |
PREDICTED: uncharacterized protein LOC105036273 [Elaeis guineensis] |
| 8 |
Hb_001009_310 |
0.259438978 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002686_290 |
0.2595220583 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15010, mitochondrial [Populus euphratica] |
| 10 |
Hb_003786_020 |
0.2626288185 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 11 |
Hb_000479_130 |
0.2649191728 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350 [Jatropha curcas] |
| 12 |
Hb_001006_230 |
0.2659716069 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_006371_010 |
0.2713885646 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 14 |
Hb_000062_150 |
0.2715157346 |
- |
- |
PREDICTED: probable carboxylesterase 16 [Jatropha curcas] |
| 15 |
Hb_007632_220 |
0.2724530063 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 16 |
Hb_002783_260 |
0.273082075 |
- |
- |
hypothetical protein JCGZ_08219 [Jatropha curcas] |
| 17 |
Hb_006111_070 |
0.2742106228 |
- |
- |
hypothetical protein POPTR_0008s07660g [Populus trichocarpa] |
| 18 |
Hb_007416_270 |
0.2751037611 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 19 |
Hb_004048_090 |
0.2755831955 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g53330 [Jatropha curcas] |
| 20 |
Hb_000088_010 |
0.2757201254 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |