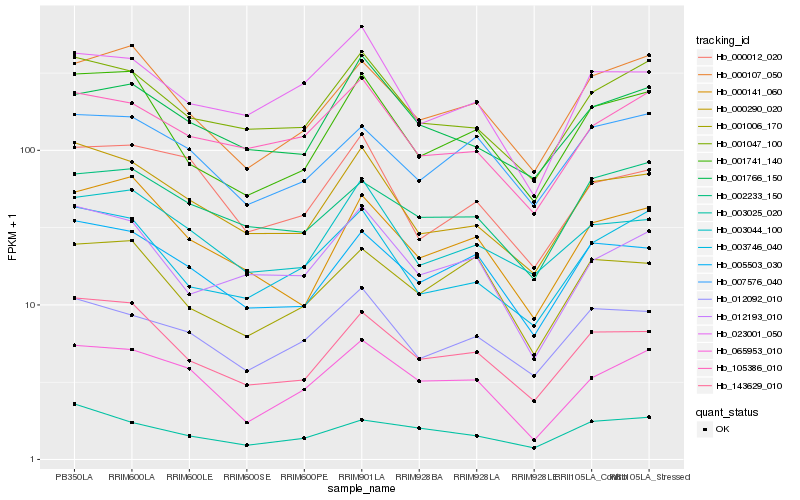
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065953_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 2 |
Hb_000107_050 |
0.1035902698 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 3 |
Hb_001047_100 |
0.1048878461 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 4 |
Hb_105386_010 |
0.1061538282 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 5 |
Hb_005503_030 |
0.1083210116 |
- |
- |
PREDICTED: protein SCO1 homolog 1, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000012_020 |
0.1088074521 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 7 |
Hb_000290_020 |
0.113283748 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K-like isoform X2 [Populus euphratica] |
| 8 |
Hb_143629_010 |
0.1147235526 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 9 |
Hb_012092_010 |
0.1165856699 |
- |
- |
PREDICTED: uncharacterized protein LOC105635929 [Jatropha curcas] |
| 10 |
Hb_001766_150 |
0.1191921072 |
- |
- |
PREDICTED: 40S ribosomal protein S6 [Jatropha curcas] |
| 11 |
Hb_007576_040 |
0.1192139887 |
- |
- |
PREDICTED: uncharacterized protein LOC105169221 [Sesamum indicum] |
| 12 |
Hb_003025_020 |
0.1218020152 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_012193_010 |
0.1218576105 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_002233_150 |
0.1223538094 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |
| 15 |
Hb_003746_040 |
0.1235163566 |
- |
- |
PREDICTED: protein N-terminal glutamine amidohydrolase [Jatropha curcas] |
| 16 |
Hb_003044_100 |
0.1235784281 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |
| 17 |
Hb_023001_050 |
0.1241939214 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 18 |
Hb_000141_060 |
0.124549084 |
- |
- |
PREDICTED: uncharacterized protein LOC105632248 [Jatropha curcas] |
| 19 |
Hb_001006_170 |
0.1249751583 |
- |
- |
Endonuclease V, putative [Ricinus communis] |
| 20 |
Hb_001741_140 |
0.1256311745 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |