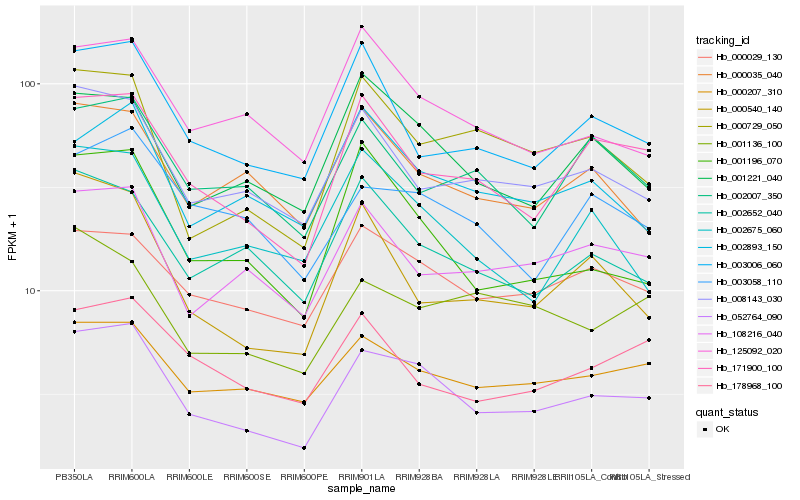
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052764_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_171900_100 |
0.1036030954 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0270496 [Jatropha curcas] |
| 3 |
Hb_000207_310 |
0.1086060907 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 4 |
Hb_000029_130 |
0.1103603182 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_008143_030 |
0.1117178551 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001221_040 |
0.1123454265 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 7 |
Hb_001196_070 |
0.1162591449 |
- |
- |
nucleolar protein nop56, putative [Ricinus communis] |
| 8 |
Hb_108216_040 |
0.1167735963 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 2 [Jatropha curcas] |
| 9 |
Hb_000729_050 |
0.1172942217 |
- |
- |
Pinin, putative [Ricinus communis] |
| 10 |
Hb_003006_060 |
0.1204387904 |
- |
- |
PREDICTED: pre-mRNA-splicing factor syf2 [Jatropha curcas] |
| 11 |
Hb_003058_110 |
0.1213713755 |
- |
- |
PREDICTED: alpha-mannosidase isoform X2 [Populus euphratica] |
| 12 |
Hb_002652_040 |
0.1216372313 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 13 |
Hb_125092_020 |
0.122293591 |
- |
- |
PREDICTED: nucleolin 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002675_060 |
0.1259610077 |
- |
- |
PREDICTED: uncharacterized protein LOC105634954 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002893_150 |
0.1261234586 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 16 |
Hb_000035_040 |
0.1265248238 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 17 |
Hb_000540_140 |
0.1306797622 |
- |
- |
zinc binding dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_001136_100 |
0.1311499324 |
- |
- |
hypothetical protein MTR_085s0013 [Medicago truncatula] |
| 19 |
Hb_178968_100 |
0.1315519692 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |
| 20 |
Hb_002007_350 |
0.1323548611 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |