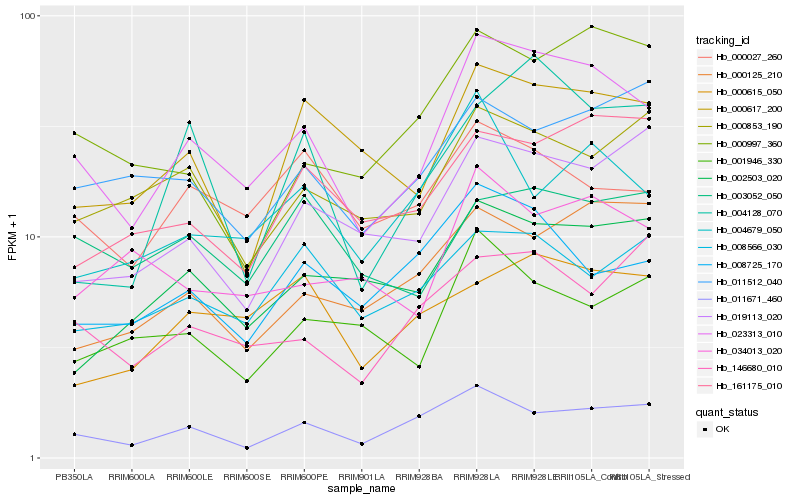
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023313_010 |
0.0 |
- |
- |
PREDICTED: linolenate hydroperoxide lyase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_011671_460 |
0.1406799439 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 3 |
Hb_000617_200 |
0.1492596399 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 4 |
Hb_008725_170 |
0.1543092674 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |
| 5 |
Hb_000027_260 |
0.1582319604 |
- |
- |
PREDICTED: chaperone protein dnaJ 15 [Jatropha curcas] |
| 6 |
Hb_034013_020 |
0.1582971164 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 7 |
Hb_002503_020 |
0.1607700422 |
- |
- |
Phosphatase 2C family protein, putative [Theobroma cacao] |
| 8 |
Hb_000853_190 |
0.1618410946 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 9 |
Hb_019113_020 |
0.1635711503 |
- |
- |
hypothetical protein JCGZ_13398 [Jatropha curcas] |
| 10 |
Hb_000997_360 |
0.1645065735 |
- |
- |
hypothetical protein AALP_AAs70636U000100 [Arabis alpina] |
| 11 |
Hb_004679_050 |
0.1648741743 |
- |
- |
galactono-1,4-lactone dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_161175_010 |
0.1648865635 |
- |
- |
PREDICTED: vesicle-associated protein 1-1 [Jatropha curcas] |
| 13 |
Hb_000125_210 |
0.1664912924 |
- |
- |
PREDICTED: electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_146680_010 |
0.1672787968 |
- |
- |
hypothetical protein JCGZ_15486 [Jatropha curcas] |
| 15 |
Hb_001946_330 |
0.16730316 |
- |
- |
PREDICTED: transmembrane protein 128 [Jatropha curcas] |
| 16 |
Hb_003052_050 |
0.1681458181 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 17 |
Hb_008566_030 |
0.1690599294 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 18 |
Hb_004128_070 |
0.1696898435 |
- |
- |
PREDICTED: thioredoxin-like protein AAED1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_011512_040 |
0.1700707249 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 20 |
Hb_000615_050 |
0.1701986484 |
- |
- |
Potassium transporter 11 family protein [Populus trichocarpa] |