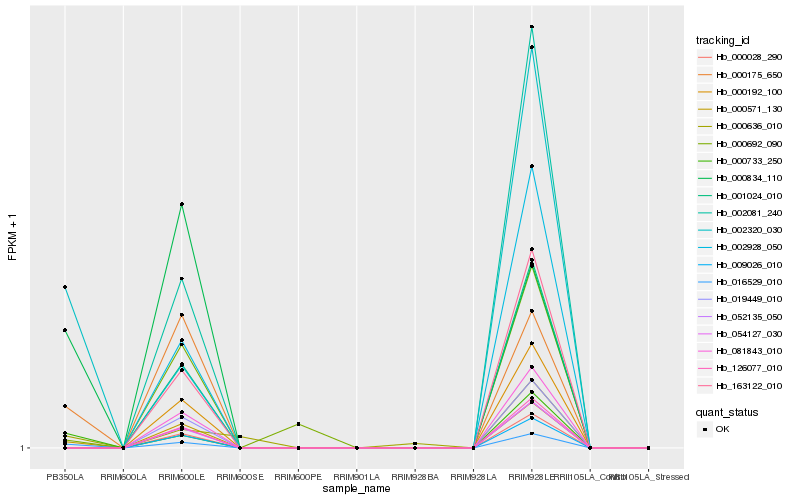
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016529_010 |
0.0 |
- |
- |
polyprotein [Oryza australiensis] |
| 2 |
Hb_000733_250 |
0.0952970429 |
- |
- |
serine/threonine protein phosphatase PP1 isozyme 1 [Loa loa] |
| 3 |
Hb_000571_130 |
0.1165325041 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_002320_030 |
0.1513683866 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000175_650 |
0.1768681309 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 6 |
Hb_000834_110 |
0.2409366664 |
- |
- |
uncharacterized protein LOC100501873 [Zea mays] |
| 7 |
Hb_000636_010 |
0.2778263936 |
- |
- |
PREDICTED: uncharacterized protein LOC105767086 [Gossypium raimondii] |
| 8 |
Hb_000692_090 |
0.2831425175 |
- |
- |
- |
| 9 |
Hb_126077_010 |
0.2908489245 |
- |
- |
- |
| 10 |
Hb_001024_010 |
0.2908831931 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 11 |
Hb_000028_290 |
0.2908856679 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 12 |
Hb_052135_050 |
0.2910059498 |
- |
- |
PREDICTED: probable pectinesterase 67 [Jatropha curcas] |
| 13 |
Hb_009026_010 |
0.2910349684 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 14 |
Hb_163122_010 |
0.2911842272 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_081843_010 |
0.2912070187 |
- |
- |
PREDICTED: uncharacterized protein LOC104880938 [Vitis vinifera] |
| 16 |
Hb_054127_030 |
0.2912699095 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 17 |
Hb_000192_100 |
0.2916816346 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 18 |
Hb_019449_010 |
0.2917155783 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_002928_050 |
0.2921738982 |
- |
- |
- |
| 20 |
Hb_002081_240 |
0.2925319637 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |