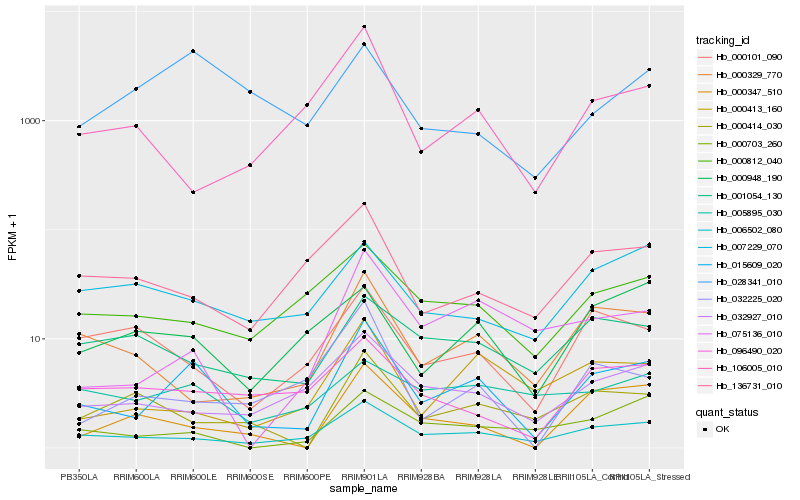
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015609_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642775 [Jatropha curcas] |
| 2 |
Hb_006502_080 |
0.2041838053 |
- |
- |
- |
| 3 |
Hb_032927_010 |
0.247687683 |
- |
- |
- |
| 4 |
Hb_000347_510 |
0.2526386132 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 5 |
Hb_075136_010 |
0.2532010275 |
- |
- |
Protein SSU72, putative [Ricinus communis] |
| 6 |
Hb_000413_160 |
0.2556395789 |
- |
- |
PREDICTED: uncharacterized protein LOC105649406 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000329_770 |
0.2562253088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000812_040 |
0.257357597 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 9 |
Hb_000703_260 |
0.257707548 |
- |
- |
N-acetylglucosamine kinase, putative [Ricinus communis] |
| 10 |
Hb_096490_020 |
0.2615529259 |
- |
- |
PREDICTED: UPF0565 protein C2orf69 [Jatropha curcas] |
| 11 |
Hb_000948_190 |
0.2619926271 |
transcription factor |
TF Family: GNAT |
Glucosamine 6-phosphate N-acetyltransferase, putative [Ricinus communis] |
| 12 |
Hb_136731_010 |
0.2655993755 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X4 [Jatropha curcas] |
| 13 |
Hb_032225_020 |
0.2677219438 |
- |
- |
hypothetical protein POPTR_0006s08140g [Populus trichocarpa] |
| 14 |
Hb_000101_090 |
0.268966307 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Camelina sativa] |
| 15 |
Hb_000414_030 |
0.2698757025 |
- |
- |
- |
| 16 |
Hb_028341_010 |
0.2708496727 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 17 |
Hb_007229_070 |
0.2719536863 |
- |
- |
PREDICTED: UPF0565 protein C2orf69 [Jatropha curcas] |
| 18 |
Hb_106005_010 |
0.2754735014 |
- |
- |
EG5651 [Manihot esculenta] |
| 19 |
Hb_001054_130 |
0.2755501835 |
- |
- |
Protein SSU72, putative [Ricinus communis] |
| 20 |
Hb_005895_030 |
0.2768635437 |
- |
- |
PREDICTED: uncharacterized protein LOC105643388 [Jatropha curcas] |