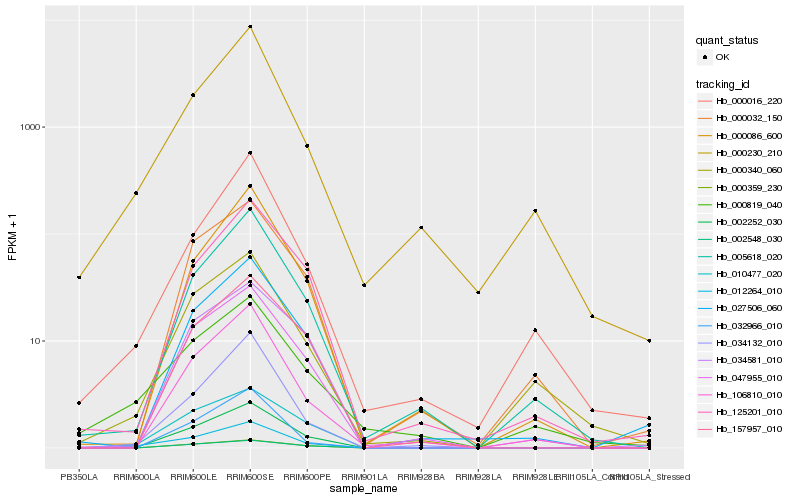
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012264_010 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002252_030 |
0.1118255434 |
- |
- |
hypothetical protein JCGZ_06771 [Jatropha curcas] |
| 3 |
Hb_010477_020 |
0.120188881 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 4 |
Hb_034132_010 |
0.1523820758 |
- |
- |
hypothetical protein B456_007G015400 [Gossypium raimondii] |
| 5 |
Hb_000819_040 |
0.1556752774 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000230_210 |
0.1600769341 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 7 |
Hb_047955_010 |
0.1619699289 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 8 |
Hb_005618_020 |
0.1648973306 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 9 |
Hb_000340_060 |
0.1679316181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_106810_010 |
0.1715148832 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_157957_010 |
0.1717144127 |
- |
- |
hypothetical protein JCGZ_10722 [Jatropha curcas] |
| 12 |
Hb_027506_060 |
0.1719993177 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 13 |
Hb_000359_230 |
0.1720712905 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 14 |
Hb_002548_030 |
0.1734018769 |
- |
- |
PREDICTED: uncharacterized protein LOC103343650 [Prunus mume] |
| 15 |
Hb_000086_600 |
0.1736303033 |
- |
- |
- |
| 16 |
Hb_032966_010 |
0.1747141341 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 17 |
Hb_000032_150 |
0.175323041 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1691430 [Ricinus communis] |
| 18 |
Hb_125201_010 |
0.1754665127 |
- |
- |
hypothetical protein POPTR_0004s03690g [Populus trichocarpa] |
| 19 |
Hb_034581_010 |
0.1772460157 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 20 |
Hb_000016_220 |
0.1783944159 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |