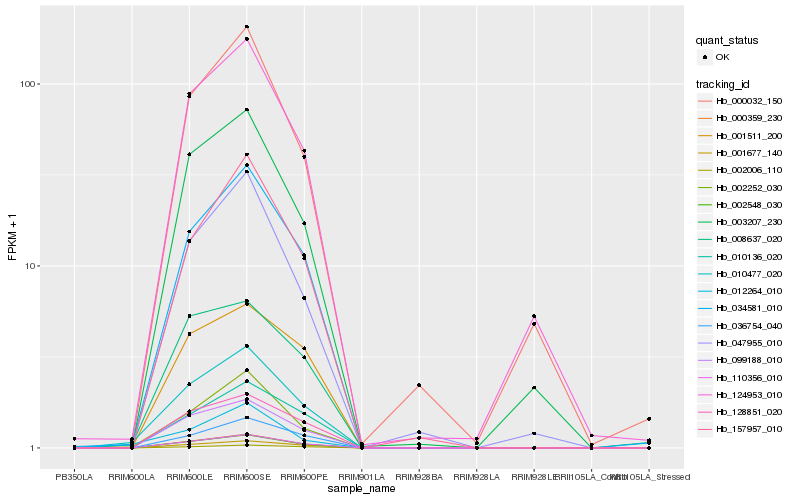
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010477_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 2 |
Hb_002548_030 |
0.0882174328 |
- |
- |
PREDICTED: uncharacterized protein LOC103343650 [Prunus mume] |
| 3 |
Hb_000359_230 |
0.0883347944 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 4 |
Hb_034581_010 |
0.0886726839 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 5 |
Hb_110356_010 |
0.088943913 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 6 |
Hb_002252_030 |
0.0917265171 |
- |
- |
hypothetical protein JCGZ_06771 [Jatropha curcas] |
| 7 |
Hb_099188_010 |
0.0986789146 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_001677_140 |
0.1036091801 |
- |
- |
PREDICTED: uncharacterized protein LOC102622520 isoform X1 [Citrus sinensis] |
| 9 |
Hb_128851_020 |
0.1101430269 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_157957_010 |
0.1170867616 |
- |
- |
hypothetical protein JCGZ_10722 [Jatropha curcas] |
| 11 |
Hb_036754_040 |
0.1179900658 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_012264_010 |
0.120188881 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003207_230 |
0.1208244749 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 14 |
Hb_010136_020 |
0.1223548576 |
- |
- |
stress-induced receptor-like kinase [Medicago truncatula] |
| 15 |
Hb_124953_010 |
0.1229951352 |
- |
- |
hypothetical protein RCOM_1314600 [Ricinus communis] |
| 16 |
Hb_047955_010 |
0.124310502 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 17 |
Hb_002006_110 |
0.1244873596 |
transcription factor |
TF Family: LFY |
LEAFY-like protein [Pistacia chinensis] |
| 18 |
Hb_008637_020 |
0.1282483622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001511_200 |
0.1308871565 |
- |
- |
Embryonic abundant protein, putative [Ricinus communis] |
| 20 |
Hb_000032_150 |
0.1350895597 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1691430 [Ricinus communis] |