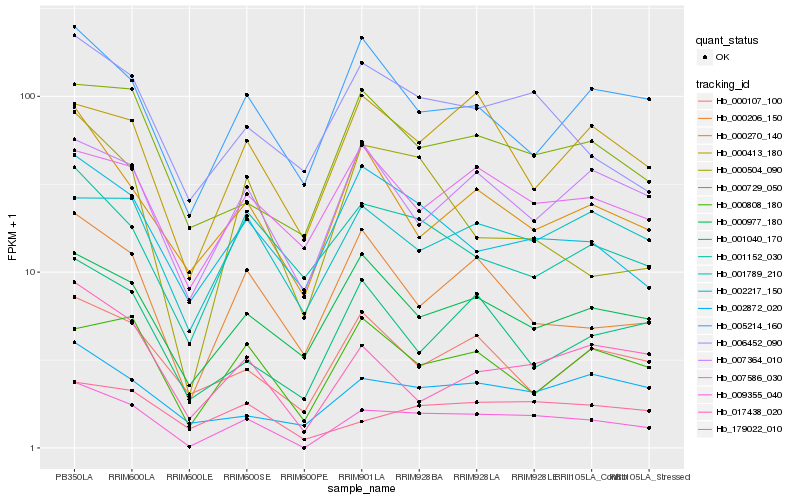
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009355_040 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_017438_020 |
0.1813081336 |
- |
- |
- |
| 3 |
Hb_179022_010 |
0.1870741561 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_006452_090 |
0.1877493444 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 5B [Jatropha curcas] |
| 5 |
Hb_002872_020 |
0.1878211021 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_007364_010 |
0.1890743657 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000206_150 |
0.1909301048 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_000729_050 |
0.1918073272 |
- |
- |
Pinin, putative [Ricinus communis] |
| 9 |
Hb_000977_180 |
0.1926838893 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001789_210 |
0.1927970609 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000270_140 |
0.1951832762 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002217_150 |
0.1957775502 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 13 |
Hb_000808_180 |
0.1965004711 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09190 [Jatropha curcas] |
| 14 |
Hb_000504_090 |
0.1983070099 |
- |
- |
PREDICTED: titin isoform X10 [Jatropha curcas] |
| 15 |
Hb_005214_160 |
0.1991755745 |
- |
- |
PREDICTED: heat shock 70 kDa protein, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000413_180 |
0.2024535934 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 17 |
Hb_007586_030 |
0.202909421 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001152_030 |
0.2031384583 |
- |
- |
hypothetical protein POPTR_0002s22050g [Populus trichocarpa] |
| 19 |
Hb_000107_100 |
0.2039774096 |
- |
- |
hypothetical protein JCGZ_16208 [Jatropha curcas] |
| 20 |
Hb_001040_170 |
0.2040364683 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |