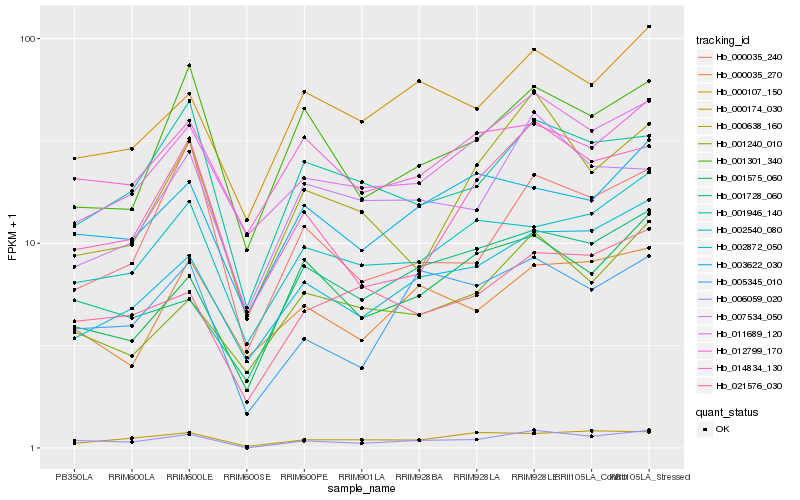
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006059_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648672 [Jatropha curcas] |
| 2 |
Hb_005345_010 |
0.1274626378 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 3 |
Hb_021576_030 |
0.1424151993 |
- |
- |
PREDICTED: uncharacterized protein LOC105637542 [Jatropha curcas] |
| 4 |
Hb_002872_050 |
0.1482639574 |
- |
- |
PREDICTED: thioredoxin-like 3-2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_014834_130 |
0.1502035867 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_001946_140 |
0.1509067008 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_011689_120 |
0.1514552834 |
- |
- |
PREDICTED: uncharacterized protein At4g15545 [Jatropha curcas] |
| 8 |
Hb_002540_080 |
0.1519787068 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |
| 9 |
Hb_000107_150 |
0.1520594251 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 10 |
Hb_001575_060 |
0.1526706962 |
- |
- |
PREDICTED: 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_003622_030 |
0.1555399803 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 1 [Jatropha curcas] |
| 12 |
Hb_000174_030 |
0.157879118 |
- |
- |
hypothetical protein RCOM_1077370 [Ricinus communis] |
| 13 |
Hb_000035_240 |
0.1588150496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001240_010 |
0.1620900143 |
- |
- |
PREDICTED: uncharacterized protein LOC105634101 [Jatropha curcas] |
| 15 |
Hb_001728_060 |
0.1624686953 |
- |
- |
PREDICTED: uncharacterized protein LOC105632289 [Jatropha curcas] |
| 16 |
Hb_001301_340 |
0.1627472631 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 17 |
Hb_000638_160 |
0.163593328 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 18 |
Hb_012799_170 |
0.1638483654 |
- |
- |
PREDICTED: protein GLC8 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007534_050 |
0.1638677148 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000035_270 |
0.1639702302 |
- |
- |
PREDICTED: uncharacterized protein LOC105637542 [Jatropha curcas] |