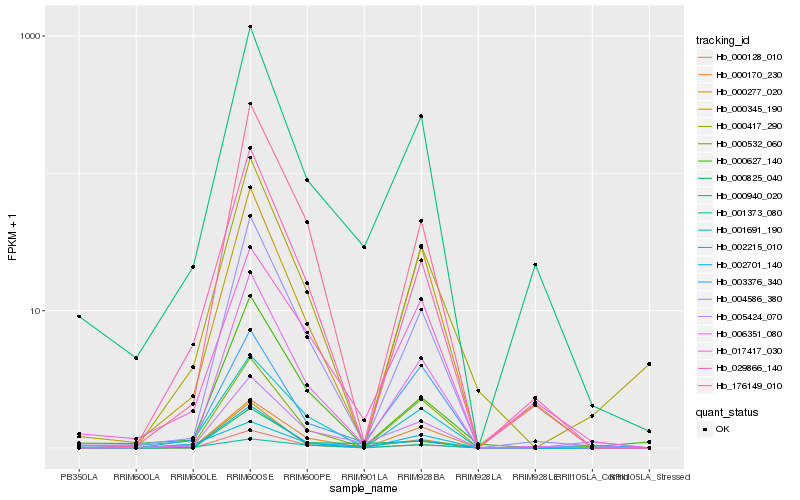
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005424_070 |
0.0 |
- |
- |
PREDICTED: TATA-box-binding protein 1-like [Jatropha curcas] |
| 2 |
Hb_000532_060 |
0.1695412172 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 3 |
Hb_004586_380 |
0.1714372102 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 4 |
Hb_000940_020 |
0.1718125198 |
- |
- |
hypothetical protein POPTR_0004s17500g [Populus trichocarpa] |
| 5 |
Hb_006351_080 |
0.1738430676 |
- |
- |
hypothetical protein B456_011G275400 [Gossypium raimondii] |
| 6 |
Hb_001691_190 |
0.1784359422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002701_140 |
0.183275704 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 8 |
Hb_000170_230 |
0.1858749607 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001373_080 |
0.1859864846 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000417_290 |
0.1913527576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_017417_030 |
0.1932463645 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 12 |
Hb_176149_010 |
0.1936950776 |
- |
- |
hypothetical protein JCGZ_16793 [Jatropha curcas] |
| 13 |
Hb_000128_010 |
0.1939188088 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 14 |
Hb_000825_040 |
0.1946982003 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_029866_140 |
0.1990194329 |
- |
- |
PREDICTED: hevamine-A [Jatropha curcas] |
| 16 |
Hb_000277_020 |
0.1992110845 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 17 |
Hb_002215_010 |
0.1994510278 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_003376_340 |
0.2006230452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000345_190 |
0.2022700454 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |
| 20 |
Hb_000627_140 |
0.2066329419 |
- |
- |
ATP binding protein, putative [Ricinus communis] |