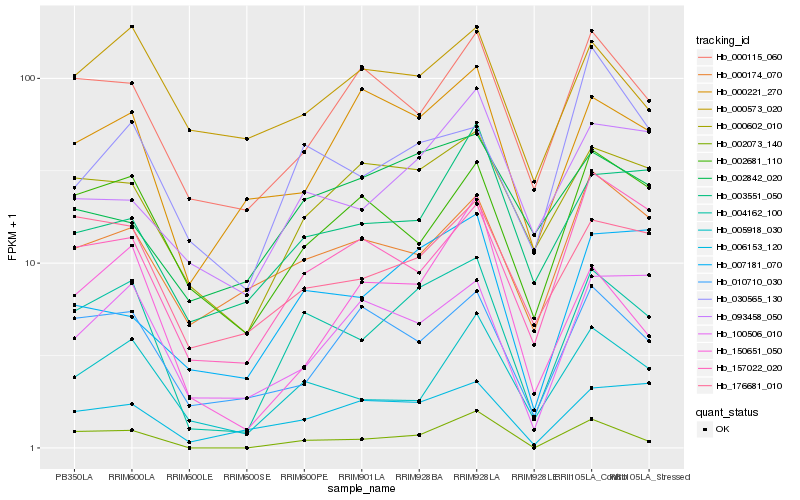
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004162_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005918_030 |
0.1660222813 |
- |
- |
hypothetical protein JCGZ_24769 [Jatropha curcas] |
| 3 |
Hb_176681_010 |
0.1727450205 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 4 |
Hb_000602_010 |
0.1731128217 |
- |
- |
PREDICTED: rhomboid-like protease 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002073_140 |
0.1815240556 |
- |
- |
- |
| 6 |
Hb_002681_110 |
0.1850757591 |
- |
- |
PREDICTED: uncharacterized protein LOC105633759 [Jatropha curcas] |
| 7 |
Hb_157022_020 |
0.1878894777 |
- |
- |
nicotinamide mononucleotide adenylyltransferase, putative [Ricinus communis] |
| 8 |
Hb_006153_120 |
0.1881024864 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 9 |
Hb_010710_030 |
0.1945811064 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription factor LUX [Jatropha curcas] |
| 10 |
Hb_000115_060 |
0.1951757091 |
- |
- |
PREDICTED: uncharacterized protein LOC103341483 [Prunus mume] |
| 11 |
Hb_007181_070 |
0.1962362766 |
- |
- |
PREDICTED: alpha-galactosidase-like [Jatropha curcas] |
| 12 |
Hb_000174_070 |
0.1996334563 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000221_270 |
0.1999931276 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 14 |
Hb_150651_050 |
0.2000329557 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 15 |
Hb_093458_050 |
0.2017281069 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 16 |
Hb_100506_010 |
0.2018876372 |
- |
- |
PREDICTED: uncharacterized protein LOC105133651 isoform X2 [Populus euphratica] |
| 17 |
Hb_030565_130 |
0.202106149 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 18 |
Hb_003551_050 |
0.2038563369 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 19 |
Hb_000573_020 |
0.2048379889 |
transcription factor |
TF Family: Orphans |
receptor histidine kinase, putative [Ricinus communis] |
| 20 |
Hb_002842_020 |
0.2053482811 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |