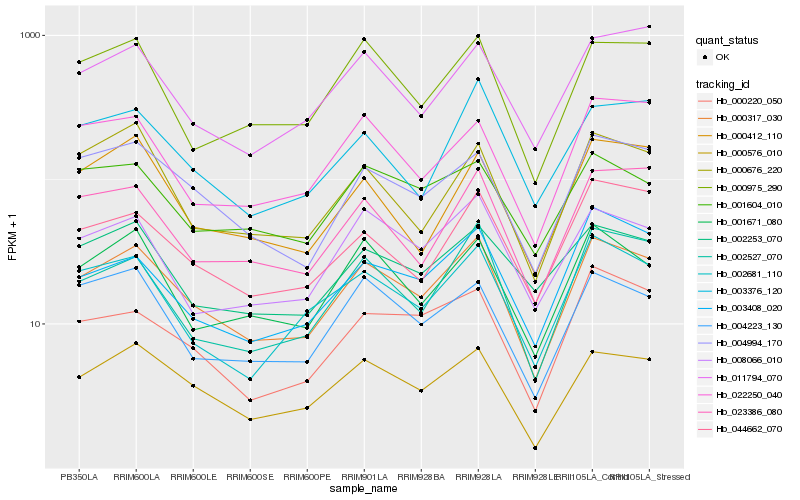
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_030 |
0.0 |
transcription factor |
TF Family: ULT |
PREDICTED: protein ULTRAPETALA 1-like [Jatropha curcas] |
| 2 |
Hb_000576_010 |
0.0597577008 |
- |
- |
PREDICTED: uncharacterized protein LOC105638289 [Jatropha curcas] |
| 3 |
Hb_004994_170 |
0.0807136867 |
- |
- |
PREDICTED: uncharacterized protein LOC105643024 [Jatropha curcas] |
| 4 |
Hb_001671_080 |
0.0897626263 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_002681_110 |
0.0920495862 |
- |
- |
PREDICTED: uncharacterized protein LOC105633759 [Jatropha curcas] |
| 6 |
Hb_004223_130 |
0.0924743036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002527_070 |
0.0927630165 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 8 |
Hb_008066_010 |
0.094448202 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit alpha-like [Jatropha curcas] |
| 9 |
Hb_000975_290 |
0.0947247839 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 10 |
Hb_023386_080 |
0.0974853938 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 11 |
Hb_044662_070 |
0.0995798353 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3 [Jatropha curcas] |
| 12 |
Hb_000676_220 |
0.101927991 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 13 |
Hb_001604_010 |
0.1036140045 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 14 |
Hb_000220_050 |
0.1041407714 |
transcription factor |
TF Family: Orphans |
PREDICTED: putative zinc finger protein At1g68190 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011794_070 |
0.1044043068 |
- |
- |
PREDICTED: 60S ribosomal protein L36-2-like [Jatropha curcas] |
| 16 |
Hb_002253_070 |
0.1052505922 |
transcription factor |
TF Family: NAC |
NAC transcription factor 042 [Jatropha curcas] |
| 17 |
Hb_022250_040 |
0.1053393595 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_003408_020 |
0.1058679334 |
- |
- |
hypothetical protein RCOM_0453040 [Ricinus communis] |
| 19 |
Hb_000412_110 |
0.1075607986 |
- |
- |
Rop4 [Hevea brasiliensis] |
| 20 |
Hb_003376_120 |
0.1075994609 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |