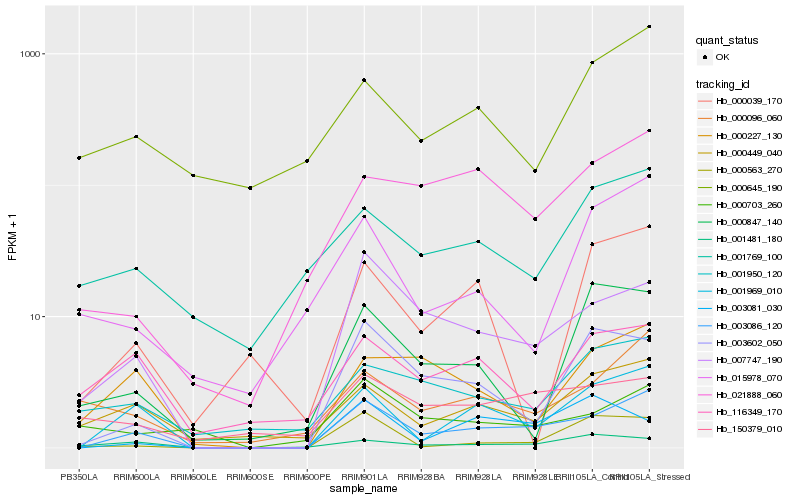
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003086_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628509 [Jatropha curcas] |
| 2 |
Hb_001969_010 |
0.1755103858 |
- |
- |
PREDICTED: probable protein Pop3 [Nelumbo nucifera] |
| 3 |
Hb_007747_190 |
0.1920648894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003602_050 |
0.2377000501 |
- |
- |
zinc binding dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_000449_040 |
0.2467090116 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_000227_130 |
0.2504565264 |
- |
- |
PREDICTED: peroxiredoxin-2F, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000096_060 |
0.2549683597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_021888_060 |
0.2550238663 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 15 homolog [Jatropha curcas] |
| 9 |
Hb_000563_270 |
0.2631064343 |
- |
- |
- |
| 10 |
Hb_001950_120 |
0.2684563568 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000703_260 |
0.277356856 |
- |
- |
N-acetylglucosamine kinase, putative [Ricinus communis] |
| 12 |
Hb_150379_010 |
0.2778769864 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 13 |
Hb_001481_180 |
0.2797684012 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 14 |
Hb_001769_100 |
0.2808632011 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 15 |
Hb_116349_170 |
0.2843433343 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 16 |
Hb_015978_070 |
0.2846925148 |
- |
- |
- |
| 17 |
Hb_003081_030 |
0.2855200105 |
- |
- |
PREDICTED: uncharacterized protein LOC101256381 [Solanum lycopersicum] |
| 18 |
Hb_000645_190 |
0.2883434801 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 19 |
Hb_000847_140 |
0.2919631333 |
- |
- |
- |
| 20 |
Hb_000039_170 |
0.2924605185 |
- |
- |
protein MOTHER of FT and TF 1-like [Jatropha curcas] |