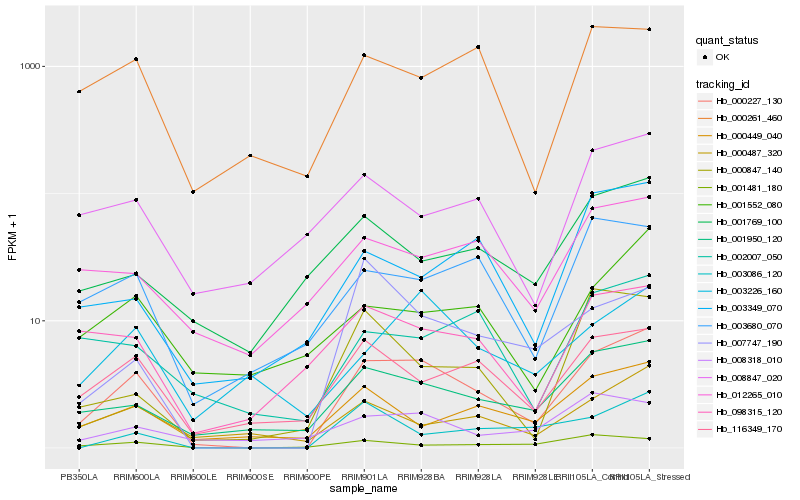
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_130 |
0.0 |
- |
- |
PREDICTED: peroxiredoxin-2F, mitochondrial [Jatropha curcas] |
| 2 |
Hb_001950_120 |
0.1939391015 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_116349_170 |
0.2074159088 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 4 |
Hb_003680_070 |
0.2268533522 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000487_320 |
0.2273257328 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 6 |
Hb_000449_040 |
0.230577098 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_003226_160 |
0.2318346869 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2-like isoform X2 [Populus euphratica] |
| 8 |
Hb_098315_120 |
0.2329082294 |
- |
- |
- |
| 9 |
Hb_000261_460 |
0.2350828273 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 10 |
Hb_001481_180 |
0.2370654622 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 11 |
Hb_002007_050 |
0.2372701179 |
- |
- |
PREDICTED: putative protease Do-like 14 [Jatropha curcas] |
| 12 |
Hb_001552_080 |
0.2399459863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012265_010 |
0.2402775599 |
- |
- |
hypothetical protein CISIN_1g026883mg [Citrus sinensis] |
| 14 |
Hb_007747_190 |
0.2436104696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003349_070 |
0.2458533981 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 16 |
Hb_008847_020 |
0.2459754574 |
- |
- |
40S ribosomal protein S17C [Hevea brasiliensis] |
| 17 |
Hb_000847_140 |
0.2469646394 |
- |
- |
- |
| 18 |
Hb_001769_100 |
0.2477912936 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 19 |
Hb_008318_010 |
0.2496639811 |
- |
- |
PREDICTED: WD repeat-containing protein 82-B isoform X1 [Jatropha curcas] |
| 20 |
Hb_003086_120 |
0.2504565264 |
- |
- |
PREDICTED: uncharacterized protein LOC105628509 [Jatropha curcas] |