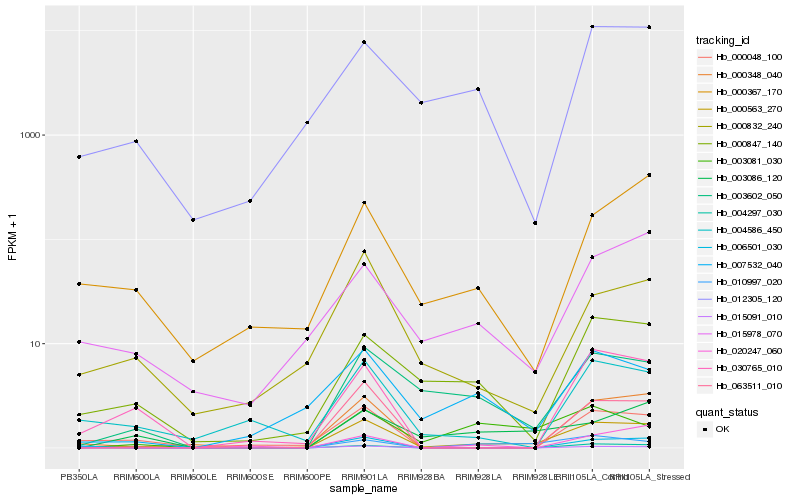
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_270 |
0.0 |
- |
- |
- |
| 2 |
Hb_003602_050 |
0.2140294551 |
- |
- |
zinc binding dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000348_040 |
0.2347379362 |
- |
- |
- |
| 4 |
Hb_000847_140 |
0.2355206606 |
- |
- |
- |
| 5 |
Hb_015091_010 |
0.2382249391 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 6 |
Hb_010997_020 |
0.2444240222 |
- |
- |
- |
| 7 |
Hb_063511_010 |
0.2476531835 |
- |
- |
DNA-directed RNA polymerase II subunit RPB4 [Gossypium arboreum] |
| 8 |
Hb_007532_040 |
0.2486994197 |
- |
- |
- |
| 9 |
Hb_006501_030 |
0.2509121751 |
- |
- |
hypothetical protein POPTR_0006s17720g [Populus trichocarpa] |
| 10 |
Hb_004297_030 |
0.2570879243 |
- |
- |
PREDICTED: uncharacterized protein LOC105633198 [Jatropha curcas] |
| 11 |
Hb_015978_070 |
0.257371975 |
- |
- |
- |
| 12 |
Hb_000832_240 |
0.2575392326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012305_120 |
0.2587012194 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 14 |
Hb_004586_450 |
0.2620224784 |
- |
- |
- |
| 15 |
Hb_003086_120 |
0.2631064343 |
- |
- |
PREDICTED: uncharacterized protein LOC105628509 [Jatropha curcas] |
| 16 |
Hb_030765_010 |
0.2638239298 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter G family member 11-like isoform X1 [Solanum lycopersicum] |
| 17 |
Hb_000048_100 |
0.2651010927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003081_030 |
0.2663310104 |
- |
- |
PREDICTED: uncharacterized protein LOC101256381 [Solanum lycopersicum] |
| 19 |
Hb_000367_170 |
0.266369191 |
- |
- |
PREDICTED: LYR motif-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_020247_060 |
0.2690760701 |
- |
- |
UDP-glucoronosyl/UDP-glucosyl transferase family protein [Populus trichocarpa] |