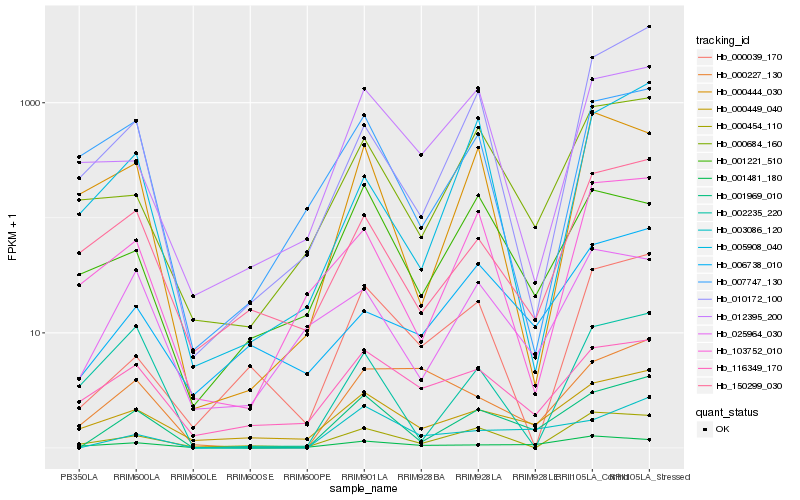
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001969_010 |
0.0 |
- |
- |
PREDICTED: probable protein Pop3 [Nelumbo nucifera] |
| 2 |
Hb_003086_120 |
0.1755103858 |
- |
- |
PREDICTED: uncharacterized protein LOC105628509 [Jatropha curcas] |
| 3 |
Hb_000449_040 |
0.2208915576 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000684_160 |
0.2298062738 |
- |
- |
PREDICTED: lipid phosphate phosphatase gamma, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000454_110 |
0.2462056286 |
- |
- |
PREDICTED: polyprenol reductase 2-like isoform X3 [Populus euphratica] |
| 6 |
Hb_150299_030 |
0.2490940752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_025964_030 |
0.250184018 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g55760 [Jatropha curcas] |
| 8 |
Hb_103752_010 |
0.2526284515 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 9 |
Hb_116349_170 |
0.2573125929 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 10 |
Hb_000039_170 |
0.2601170882 |
- |
- |
protein MOTHER of FT and TF 1-like [Jatropha curcas] |
| 11 |
Hb_001481_180 |
0.2602603079 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 12 |
Hb_012395_200 |
0.2628237841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006738_010 |
0.2635061295 |
- |
- |
PREDICTED: uncharacterized protein LOC105634993 [Jatropha curcas] |
| 14 |
Hb_001221_510 |
0.2646331893 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 15 |
Hb_010172_100 |
0.2647002161 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 16 |
Hb_002235_220 |
0.2648416543 |
- |
- |
PREDICTED: pectinesterase 31-like [Jatropha curcas] |
| 17 |
Hb_007747_130 |
0.2662374396 |
- |
- |
PREDICTED: uncharacterized protein LOC105635879 [Jatropha curcas] |
| 18 |
Hb_000444_030 |
0.2670868191 |
- |
- |
PREDICTED: expansin-A20 [Jatropha curcas] |
| 19 |
Hb_000227_130 |
0.2679611207 |
- |
- |
PREDICTED: peroxiredoxin-2F, mitochondrial [Jatropha curcas] |
| 20 |
Hb_005908_040 |
0.2682927421 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |