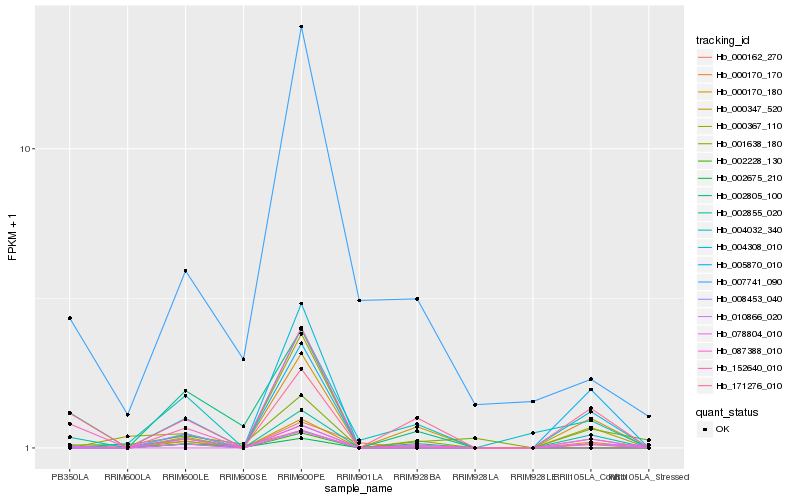
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002855_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_171276_010 |
0.131575842 |
- |
- |
- |
| 3 |
Hb_087388_010 |
0.2775874981 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 4 |
Hb_000170_180 |
0.2981652714 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 5 |
Hb_008453_040 |
0.3093405517 |
- |
- |
PREDICTED: expansin-A11 [Jatropha curcas] |
| 6 |
Hb_000347_520 |
0.311789546 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 7 |
Hb_010866_020 |
0.3214725692 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_005870_010 |
0.3245520704 |
- |
- |
- |
| 9 |
Hb_000367_110 |
0.3505117869 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |
| 10 |
Hb_002675_210 |
0.3514687682 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 11 |
Hb_004032_340 |
0.3523451477 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 12 |
Hb_002805_100 |
0.3560042424 |
- |
- |
Receptor-like protein 12 [Morus notabilis] |
| 13 |
Hb_078804_010 |
0.3579557892 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_001638_180 |
0.3591164782 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 15 |
Hb_004308_010 |
0.3607760242 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 16 |
Hb_000170_170 |
0.3623407483 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 17 |
Hb_000162_270 |
0.3630672009 |
- |
- |
PREDICTED: uncharacterized protein LOC104442671 [Eucalyptus grandis] |
| 18 |
Hb_152640_010 |
0.3640177954 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 19 |
Hb_002228_130 |
0.3656408073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007741_090 |
0.3656594159 |
- |
- |
Autophagy-related 8f -like protein [Gossypium arboreum] |