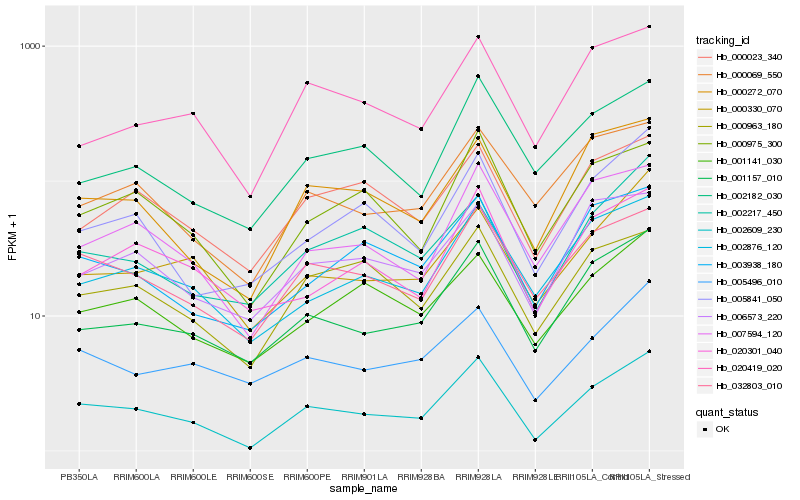
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002609_230 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 2 |
Hb_001157_010 |
0.1203893511 |
- |
- |
deoxyhypusine synthase, putative [Ricinus communis] |
| 3 |
Hb_032803_010 |
0.1243580074 |
- |
- |
PREDICTED: glycosyltransferase-like At2g41451 [Jatropha curcas] |
| 4 |
Hb_007594_120 |
0.1253012552 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 2 [Hevea brasiliensis] |
| 5 |
Hb_000272_070 |
0.1261053141 |
- |
- |
snare protein YKt [Hevea brasiliensis] |
| 6 |
Hb_000975_300 |
0.1276573167 |
- |
- |
Rab2 [Hevea brasiliensis] |
| 7 |
Hb_005841_050 |
0.1365883996 |
- |
- |
PREDICTED: F-box protein SKIP1-like [Jatropha curcas] |
| 8 |
Hb_002876_120 |
0.13677031 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase PTP1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003938_180 |
0.1405255382 |
- |
- |
lesion inducing family protein [Populus trichocarpa] |
| 10 |
Hb_002217_450 |
0.1416340828 |
- |
- |
PREDICTED: probable prefoldin subunit 3 [Jatropha curcas] |
| 11 |
Hb_000330_070 |
0.1424286194 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 12 |
Hb_000963_180 |
0.1435242856 |
- |
- |
calcium/calmodulin-dependent protein kinase kinase, putative [Ricinus communis] |
| 13 |
Hb_000069_550 |
0.1438461685 |
- |
- |
PREDICTED: uncharacterized protein LOC104596457 isoform X1 [Nelumbo nucifera] |
| 14 |
Hb_020419_020 |
0.1438550108 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 15 |
Hb_020301_040 |
0.1444973703 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein P [Jatropha curcas] |
| 16 |
Hb_002182_030 |
0.1446642191 |
- |
- |
hypothetical protein Csa_5G139175 [Cucumis sativus] |
| 17 |
Hb_005496_010 |
0.1457920638 |
- |
- |
PREDICTED: PXMP2/4 family protein 4 [Jatropha curcas] |
| 18 |
Hb_001141_030 |
0.1460907759 |
- |
- |
hypothetical protein PRUPE_ppa024854mg [Prunus persica] |
| 19 |
Hb_000023_340 |
0.1468712957 |
- |
- |
PREDICTED: enhancer of rudimentary homolog [Jatropha curcas] |
| 20 |
Hb_006573_220 |
0.1472324031 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |