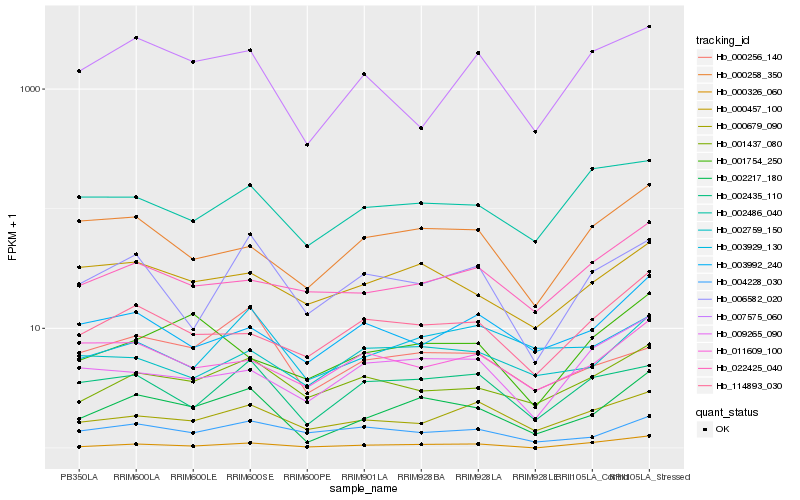
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_180 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108-like [Populus euphratica] |
| 2 |
Hb_001437_080 |
0.1738249117 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_05472 [Jatropha curcas] |
| 3 |
Hb_114893_030 |
0.1743022426 |
- |
- |
PREDICTED: ribosome-recycling factor, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002435_110 |
0.1762618363 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 5 |
Hb_000457_100 |
0.177470992 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 6 |
Hb_000258_350 |
0.182984488 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 7 |
Hb_002759_150 |
0.1861803221 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000679_090 |
0.1875489568 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 9 |
Hb_006582_020 |
0.1887553981 |
- |
- |
Zinc finger protein ZFPM1 [Theobroma cacao] |
| 10 |
Hb_009265_090 |
0.1912112723 |
- |
- |
PREDICTED: uncharacterized protein LOC105649618 [Jatropha curcas] |
| 11 |
Hb_003929_130 |
0.1933166736 |
- |
- |
PREDICTED: uncharacterized protein LOC105643628 [Jatropha curcas] |
| 12 |
Hb_004228_030 |
0.1934333567 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_001754_250 |
0.1948544261 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 14 |
Hb_003992_240 |
0.1966063196 |
- |
- |
- |
| 15 |
Hb_022425_040 |
0.1970724338 |
- |
- |
PREDICTED: probable RNA methyltransferase At5g51130 [Jatropha curcas] |
| 16 |
Hb_000326_060 |
0.1985073381 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 17 |
Hb_002486_040 |
0.1985748519 |
- |
- |
- |
| 18 |
Hb_007575_060 |
0.1987172806 |
- |
- |
hypothetical protein CARUB_v10002094mg, partial [Capsella rubella] |
| 19 |
Hb_000256_140 |
0.1994188455 |
- |
- |
PREDICTED: uncharacterized protein LOC105637585 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011609_100 |
0.2002748643 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |