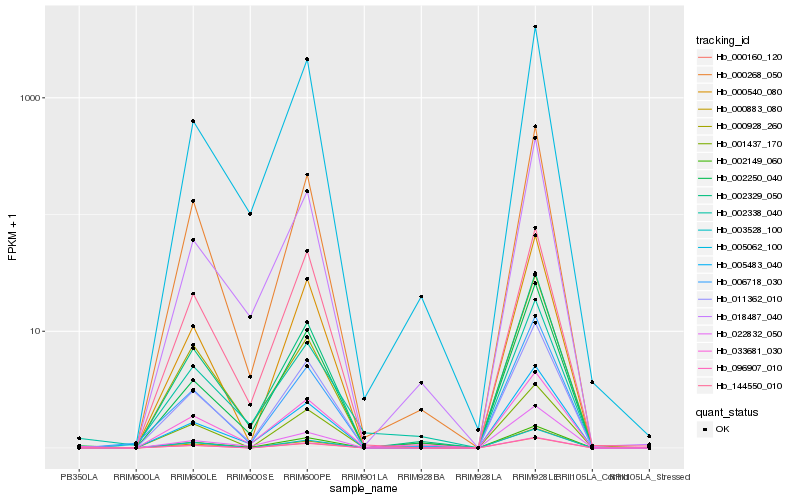
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002149_060 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_005483_040 |
0.083052325 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 3 |
Hb_011362_010 |
0.0911458588 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 4 |
Hb_002329_050 |
0.0966992703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_022832_050 |
0.0971294329 |
- |
- |
PREDICTED: protein argonaute 16 isoform X1 [Sesamum indicum] |
| 6 |
Hb_033681_030 |
0.097316341 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 7 |
Hb_000268_050 |
0.1007776564 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000883_080 |
0.1018410847 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_002338_040 |
0.1124527723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002250_040 |
0.1150713564 |
- |
- |
PREDICTED: transcription repressor OFP17 [Jatropha curcas] |
| 11 |
Hb_000160_120 |
0.1159106561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000540_080 |
0.1167944357 |
- |
- |
PREDICTED: laccase-17 [Jatropha curcas] |
| 13 |
Hb_096907_010 |
0.1170691896 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 14 |
Hb_001437_170 |
0.1171374115 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 15 |
Hb_000928_260 |
0.1174143076 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 16 |
Hb_144550_010 |
0.1188945409 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_005062_100 |
0.1190111463 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_018487_040 |
0.1195187289 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 19 |
Hb_003528_100 |
0.1198064496 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 20 |
Hb_006718_030 |
0.1203569487 |
- |
- |
ATPase, F1 complex, gamma subunit protein [Theobroma cacao] |